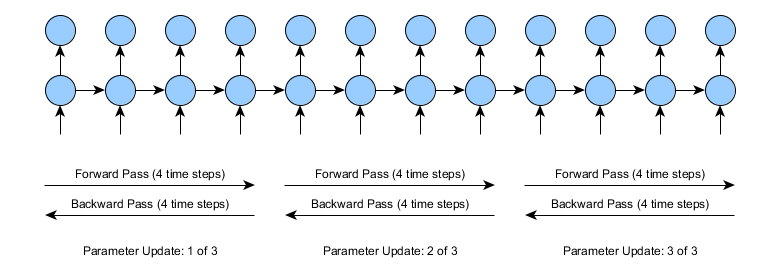
5.3 Truncated BPTT#
When the sequences are very long (thousands of points), the network training can be very slow and the memory requirements increase. The truncated BPTT is an alternative similar to mini-batch training in Dense Networks, even though in RNN the batch parameter can also be used.
!wget -nc --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/2021.deeplearning/main/content/init.py
import init; init.init(force_download=False);
from IPython.display import Image
Image(filename='local/imgs/rnn_tbptt_2.png', width=1200)
#
The TBPTT can be implemented by setting up the data appropriately. Let’s remember that for recurrent neural networks, data must have the format [n_samples,n_times,n_features], so if you want to use Truncated BPTT you just have to split the sequences into more n_samples of less n_times.
BUT, Is it possible that the LSTM may find dependencies between the sequences?#
No it’s not possible unless you go for the stateful LSTM.
So the use of Truncated BPTT requires to set up the Stateful mode.
Example#
Extracted from: http://philipperemy.github.io/keras-stateful-lstm/
Let’s see a problem of classifying sequences. The data matrix \(X\) is made exclusively of zeros except in the first column where exactly half of the values are 1.
import numpy as np
from numpy.random import choice
N_train = 1000
X_train = np.zeros((N_train,20))
one_indexes = choice(a=N_train, size=int(N_train / 2), replace=False)
X_train[one_indexes, 0] = 1 # very long term memory.
#--------------------------------
N_test = 200
X_test = np.zeros((N_test,20))
one_indexes = choice(a=N_test, size=int(N_test / 2), replace=False)
X_test[one_indexes, 0] = 1 # very long term memory.
print(X_train[:10,:5])
print(X_test[:10,:5])
[[1. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]]
[[1. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[1. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]
[0. 0. 0. 0. 0.]]
print(X_train.shape)
(1000, 20)
def prepare_sequences(x_train, y_train, window_length, increment):
windows = []
windows_y = []
for i, sequence in enumerate(x_train):
len_seq = len(sequence)
for window_start in range(0, len_seq - window_length + 1, increment):
window_end = window_start + window_length
window = sequence[window_start:window_end]
windows.append(window)
windows_y.append(y_train[i])
return np.array(windows), np.array(windows_y)
#Split the sequences into two sequences of length 10
window_length = 10
x_train, y_train = prepare_sequences(X_train, X_train[:,0], window_length,window_length)
x_test, y_test = prepare_sequences(X_test, X_test[:,0], window_length,window_length)
x_train = x_train.reshape(x_train.shape[0],x_train.shape[1],1)
x_test = x_test.reshape(x_test.shape[0],x_test.shape[1],1)
y_train = y_train.reshape(y_train.shape[0],1)
y_test = y_test.reshape(y_test.shape[0],1)
print(x_train.shape)
print(y_train.shape)
print(x_test.shape)
print(y_test.shape)
(2000, 10, 1)
(2000, 1)
(400, 10, 1)
(400, 1)
Every sequence was split into 2 subsequences
print(x_train[:4,:])
print(y_train[:4,:])
[[[1.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]]
[[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]]
[[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]]
[[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]
[0.]]]
[[1.]
[1.]
[0.]
[0.]]
Let’s train a regular LSTM network#
from tensorflow.keras.layers import LSTM, Dense, Input
from tensorflow.keras.models import Sequential
Using the original sequences:#
print('Building STATELESS model...')
max_len = 10
batch_size = 11
model = Sequential()
model.add(Input(shape=(20, 1)))
model.add(LSTM(10, return_sequences=False, stateful=False))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X_train.reshape(1000,20,1), X_train[:,0].reshape(1000,1), batch_size=batch_size, epochs=5,
validation_data=(X_test.reshape(200,20,1), X_test[:,0].reshape(200,1)), shuffle=False)
score, acc = model.evaluate(X_test.reshape(200,20,1),X_test[:,0].reshape(200,1), batch_size=batch_size, verbose=0)
Building STATELESS model...
Epoch 1/5
91/91 ━━━━━━━━━━━━━━━━━━━━ 1s 2ms/step - accuracy: 0.6195 - loss: 0.6845 - val_accuracy: 1.0000 - val_loss: 0.3576
Epoch 2/5
91/91 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 1.0000 - loss: 0.2445 - val_accuracy: 1.0000 - val_loss: 0.1088
Epoch 3/5
91/91 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 1.0000 - loss: 0.0904 - val_accuracy: 1.0000 - val_loss: 0.0507
Epoch 4/5
91/91 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 1.0000 - loss: 0.0432 - val_accuracy: 1.0000 - val_loss: 0.0281
Epoch 5/5
91/91 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 1.0000 - loss: 0.0251 - val_accuracy: 1.0000 - val_loss: 0.0185
acc
1.0
Using the splitted sequences:#
print('Building STATELESS model...')
max_len = 10
batch_size = 2
model = Sequential()
model.add(Input(shape=(max_len,1)))
model.add(LSTM(10, return_sequences=False, stateful=False))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(x_train, y_train, batch_size=batch_size, epochs=15,
validation_data=(x_test, y_test), shuffle=False)
score, acc = model.evaluate(x_test, y_test, batch_size=batch_size, verbose=0)
Building STATELESS model...
Epoch 1/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 628us/step - accuracy: 0.6097 - loss: 0.5924 - val_accuracy: 0.7500 - val_loss: 0.4794
Epoch 2/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 566us/step - accuracy: 0.7491 - loss: 0.4807 - val_accuracy: 0.7500 - val_loss: 0.4785
Epoch 3/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 570us/step - accuracy: 0.7491 - loss: 0.4798 - val_accuracy: 0.7500 - val_loss: 0.4781
Epoch 4/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 570us/step - accuracy: 0.7491 - loss: 0.4794 - val_accuracy: 0.7500 - val_loss: 0.4779
Epoch 5/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 592us/step - accuracy: 0.7491 - loss: 0.4792 - val_accuracy: 0.7500 - val_loss: 0.4778
Epoch 6/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 573us/step - accuracy: 0.7491 - loss: 0.4791 - val_accuracy: 0.7500 - val_loss: 0.4777
Epoch 7/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 574us/step - accuracy: 0.7491 - loss: 0.4789 - val_accuracy: 0.7500 - val_loss: 0.4777
Epoch 8/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 589us/step - accuracy: 0.7491 - loss: 0.4788 - val_accuracy: 0.7500 - val_loss: 0.4776
Epoch 9/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 577us/step - accuracy: 0.7491 - loss: 0.4788 - val_accuracy: 0.7500 - val_loss: 0.4776
Epoch 10/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 580us/step - accuracy: 0.7491 - loss: 0.4787 - val_accuracy: 0.7500 - val_loss: 0.4775
Epoch 11/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 581us/step - accuracy: 0.7491 - loss: 0.4786 - val_accuracy: 0.7500 - val_loss: 0.4775
Epoch 12/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 582us/step - accuracy: 0.7491 - loss: 0.4786 - val_accuracy: 0.7500 - val_loss: 0.4775
Epoch 13/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 582us/step - accuracy: 0.7491 - loss: 0.4785 - val_accuracy: 0.7500 - val_loss: 0.4775
Epoch 14/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 602us/step - accuracy: 0.7491 - loss: 0.4785 - val_accuracy: 0.7500 - val_loss: 0.4774
Epoch 15/15
1000/1000 ━━━━━━━━━━━━━━━━━━━━ 1s 582us/step - accuracy: 0.7491 - loss: 0.4785 - val_accuracy: 0.7500 - val_loss: 0.4774
acc
0.75
The sequences composed of 0s are correctly classified. The subsequences starting with 1 are correctly classified, but the sebsequences of class 1 starting with 0, are wrong classified. Those are the 25% of the sequences.
What happened?#
The long range memory required to classify the sequences correctly has been lost because of the sequences’ partition.
STATEFUL Model#
print('Build STATEFUL model...')
max_len = 10
n_partitions = 2
batch = 1
model = Sequential()
model.add(Input(shape=(max_len, 1),batch_size=1))
model.add(LSTM(10, return_sequences=False, stateful=True))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
Build STATEFUL model...
print('Train...')
for epoch in range(5):
mean_tr_acc = []
mean_tr_loss = []
for i in range(0,x_train.shape[0],n_partitions):
#print(i)
for j in range(n_partitions):
#print(j)
tr_loss, tr_acc = model.train_on_batch(x_train[i+j,:,:].reshape(1,max_len,1), y_train[i+j,:].reshape(1,1))
mean_tr_acc.append(tr_acc)
mean_tr_loss.append(tr_loss)
model.layers[0].reset_states()
print('accuracy training = {}'.format(np.mean(mean_tr_acc)))
print('loss training = {}'.format(np.mean(mean_tr_loss)))
print('___________________________________')
mean_te_acc = []
mean_te_loss = []
for i in range(0,x_test.shape[0],n_partitions):
for j in range(n_partitions):
te_loss, te_acc = model.test_on_batch(x_test[i+j,:,:].reshape(1,max_len,1), y_test[i+j,:].reshape(1,1))
mean_te_acc.append(te_acc)
mean_te_loss.append(te_loss)
model.layers[0].reset_states()
print('accuracy testing = {}'.format(np.mean(mean_te_acc)))
print('loss testing = {}'.format(np.mean(mean_te_loss)))
print('___________________________________')
Train...
accuracy training = 0.8727690577507019
loss training = 0.23629359900951385
___________________________________
accuracy testing = 0.9667946100234985
loss testing = 0.07307052612304688
___________________________________
accuracy training = 0.977907121181488
loss training = 0.048978641629219055
___________________________________
accuracy testing = 0.9841359853744507
loss testing = 0.03531878814101219
___________________________________
accuracy training = 0.9872968792915344
loss training = 0.02832924574613571
___________________________________
accuracy testing = 0.9895753264427185
loss testing = 0.023273712024092674
___________________________________
accuracy training = 0.9910579919815063
loss training = 0.019974345341324806
___________________________________
accuracy testing = 0.9922366142272949
loss testing = 0.017347801476716995
___________________________________
accuracy training = 0.9930955767631531
loss training = 0.015431100502610207
___________________________________
accuracy testing = 0.9938153028488159
loss testing = 0.013824365101754665
___________________________________
The code was a bit more difficult to write because we have to manually call model.reset_states() at each new sequence processed. Another method to do that is to write a callback that reset the states at each sequence like this:
from tensorflow.keras.callbacks import Callback
n_partitions = 2
class ResetStatesCallback(Callback):
def __init__(self):
self.counter = 0
def on_batch_begin(self, batch, logs={}):
if self.counter % n_partitions == 0:
self.model.layers[0].reset_states()
self.counter += 1
model = Sequential()
model.add(Input(shape=(max_len, 1),batch_size=1))
model.add(LSTM(10, return_sequences=False, stateful=True))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(x_train, y_train, epochs=5, callbacks=[ResetStatesCallback()], batch_size=1, shuffle=False)
Epoch 1/5
2000/2000 ━━━━━━━━━━━━━━━━━━━━ 1s 561us/step - accuracy: 0.8624 - loss: 0.2455
Epoch 2/5
2000/2000 ━━━━━━━━━━━━━━━━━━━━ 1s 557us/step - accuracy: 1.0000 - loss: 0.0012
Epoch 3/5
2000/2000 ━━━━━━━━━━━━━━━━━━━━ 1s 557us/step - accuracy: 1.0000 - loss: 2.6386e-04
Epoch 4/5
2000/2000 ━━━━━━━━━━━━━━━━━━━━ 1s 558us/step - accuracy: 1.0000 - loss: 8.2083e-05
Epoch 5/5
2000/2000 ━━━━━━━━━━━━━━━━━━━━ 1s 571us/step - accuracy: 1.0000 - loss: 2.8112e-05
<keras.src.callbacks.history.History at 0x1774afbe0>
When the dataset for validation have a different batchsize, the best way to solve it, is to create a new model with the new batchsize and transfer to it the weights of the trained model.
*Example: The following code does not have relation with the previous examples!
X = np.random.randn(100,4,2)
y = np.random.randn(100)
# configure network
n_batch = 3
n_epoch = 2
n_neurons = 10
# design network
model = Sequential()
model.add(Input(shape=(X.shape[1], X.shape[2]),batch_size=n_batch))
model.add(LSTM(n_neurons, stateful=True))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
# fit network
for i in range(n_epoch):
model.fit(X, y, epochs=1, batch_size=n_batch, verbose=1, shuffle=False)
model.layers[0].reset_states()
# re-define the batch size
n_batch = 1
# re-define model
new_model = Sequential()
new_model.add(Input(shape=(X.shape[1], X.shape[2]),batch_size=n_batch))
new_model.add(LSTM(n_neurons, stateful=True))
new_model.add(Dense(1))
# copy weights
old_weights = model.get_weights()
new_model.set_weights(old_weights)
# compile model
new_model.compile(loss='mean_squared_error', optimizer='adam')
34/34 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - loss: 1.2503
34/34 ━━━━━━━━━━━━━━━━━━━━ 0s 596us/step - loss: 1.2227