04.02 - DATA CLEANING#
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
Based on Kaggle House Pricing Prediction Competition#
Inspect and learn from the competition Notebooks
You must make available to this notebook the
train.csvfile from the competition data section. If running this notebook in Google Colab you must upload it in the notebook files section in Colab.
## KEEPOUTPUT
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from progressbar import progressbar as pbar
from local.lib import mlutils
%matplotlib inline
d = pd.read_csv("train.csv", index_col="Id")
d.head()
| MSSubClass | MSZoning | LotFrontage | LotArea | Street | Alley | LotShape | LandContour | Utilities | LotConfig | ... | PoolArea | PoolQC | Fence | MiscFeature | MiscVal | MoSold | YrSold | SaleType | SaleCondition | SalePrice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Id | |||||||||||||||||||||
| 1 | 60 | RL | 65.0 | 8450 | Pave | NaN | Reg | Lvl | AllPub | Inside | ... | 0 | NaN | NaN | NaN | 0 | 2 | 2008 | WD | Normal | 208500 |
| 2 | 20 | RL | 80.0 | 9600 | Pave | NaN | Reg | Lvl | AllPub | FR2 | ... | 0 | NaN | NaN | NaN | 0 | 5 | 2007 | WD | Normal | 181500 |
| 3 | 60 | RL | 68.0 | 11250 | Pave | NaN | IR1 | Lvl | AllPub | Inside | ... | 0 | NaN | NaN | NaN | 0 | 9 | 2008 | WD | Normal | 223500 |
| 4 | 70 | RL | 60.0 | 9550 | Pave | NaN | IR1 | Lvl | AllPub | Corner | ... | 0 | NaN | NaN | NaN | 0 | 2 | 2006 | WD | Abnorml | 140000 |
| 5 | 60 | RL | 84.0 | 14260 | Pave | NaN | IR1 | Lvl | AllPub | FR2 | ... | 0 | NaN | NaN | NaN | 0 | 12 | 2008 | WD | Normal | 250000 |
5 rows × 80 columns
## KEEPOUTPUT
print (d.shape)
(1460, 80)
We must repair the missing data in the following columns#
Possible repair actions:
Remove row or column
Replace value (why what?)
## KEEPOUTPUT
k = d.isna().sum()
k[k!=0]
LotFrontage 259
Alley 1369
MasVnrType 872
MasVnrArea 8
BsmtQual 37
BsmtCond 37
BsmtExposure 38
BsmtFinType1 37
BsmtFinType2 38
Electrical 1
FireplaceQu 690
GarageType 81
GarageYrBlt 81
GarageFinish 81
GarageQual 81
GarageCond 81
PoolQC 1453
Fence 1179
MiscFeature 1406
dtype: int64
Inspect and understand missing data#
## KEEPOUTPUT
def plot_missing(col, target):
def f1():
if d[col].dtype==object:
k = d[col].fillna("missing").value_counts()
sns.barplot(x=k.index, y=k.values)
else:
plt.hist(d[col].dropna().values, bins=100)
plt.title("distribution of %s"%col)
plt.grid()
def f2():
if d[col].dtype==object:
k=d[[col,target]].dropna()
for v in d[col].dropna().unique():
if sum(k[col]==v)>1:
sns.histplot(k[target][k[col]==v], kde=True,
label=v, alpha=.3);
if sum(d[col].isna())>1:
sns.histplot(d[target][d[col].isna()],
alpha=.8, kde=True,
label="missing")
plt.legend();
else:
plt.scatter(d[col], d[target], alpha=.5)
plt.xlabel(target)
plt.ylabel(col)
plt.grid()
plt.title("%s vs target"%(col))
def f3():
n = np.sum(d[col].isna())
if n>1:
sns.histplot(d[target][d[col].isna()], color="red", kde=True, alpha=.3, label="missing (%d values)"%n)
sns.histplot(d[target][~d[col].isna()], color="blue", kde=True, alpha=.3, label="ok (%d values)"%(len(d)-n))
plt.title("distribution of target wrt %s"%col)
plt.yticks([])
plt.grid()
plt.legend()
mlutils.figures_grid(3,1, [f1, f2, f3], figsize=(20,3))
## KEEPOUTPUT
for col in k[k!=0].index:
plot_missing(col, target="SalePrice")
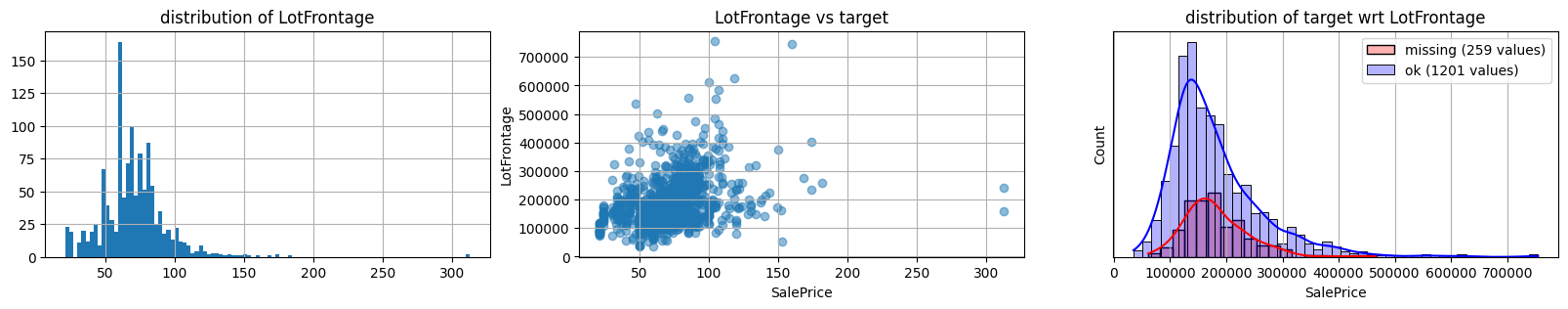
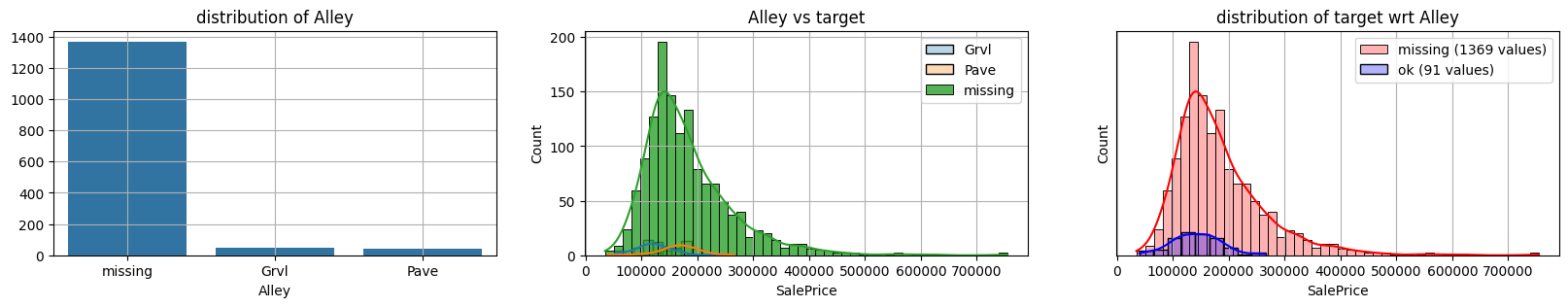
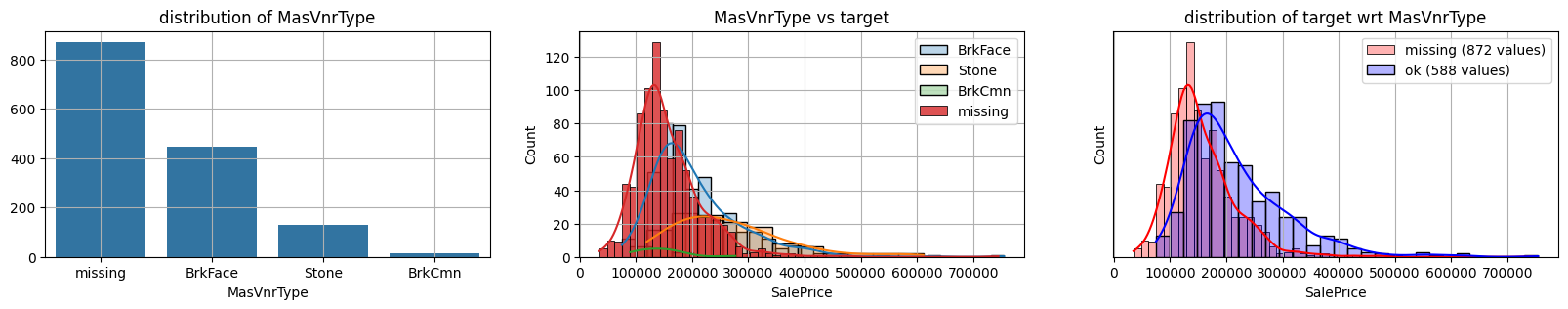
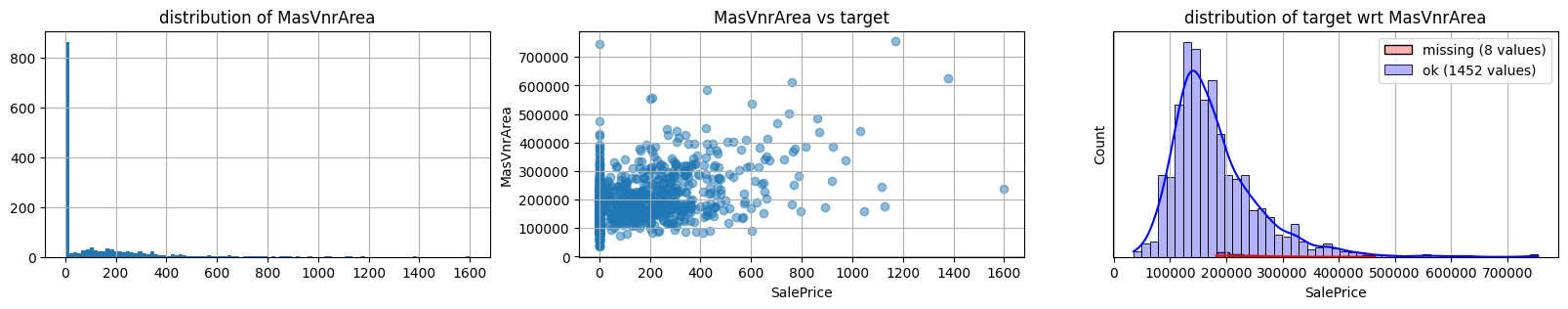
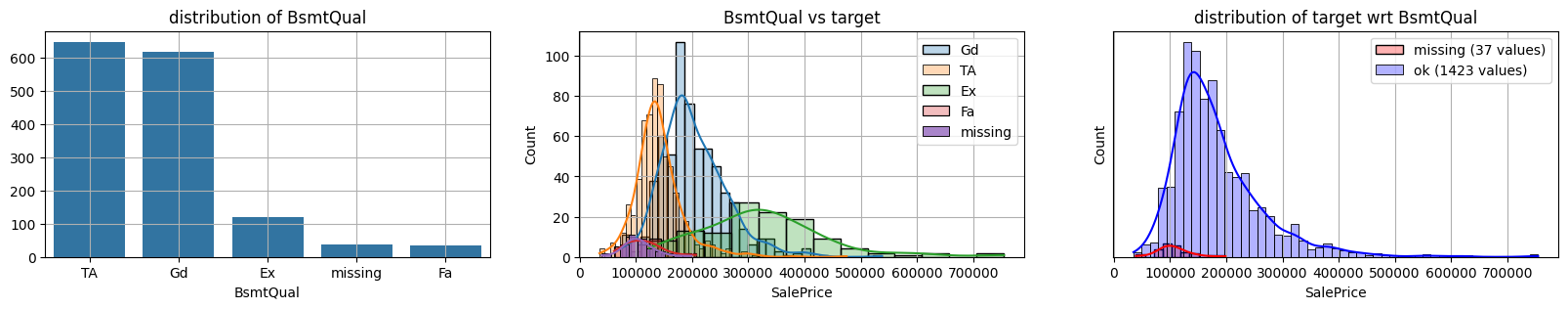
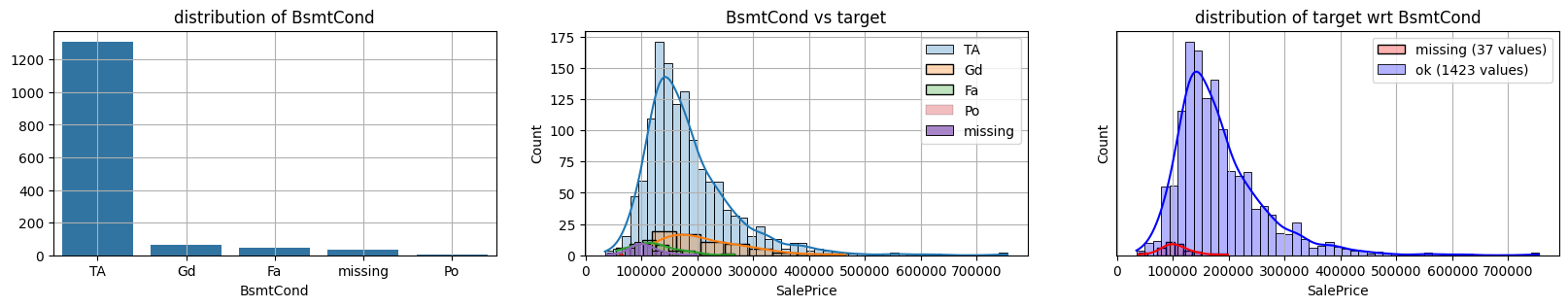
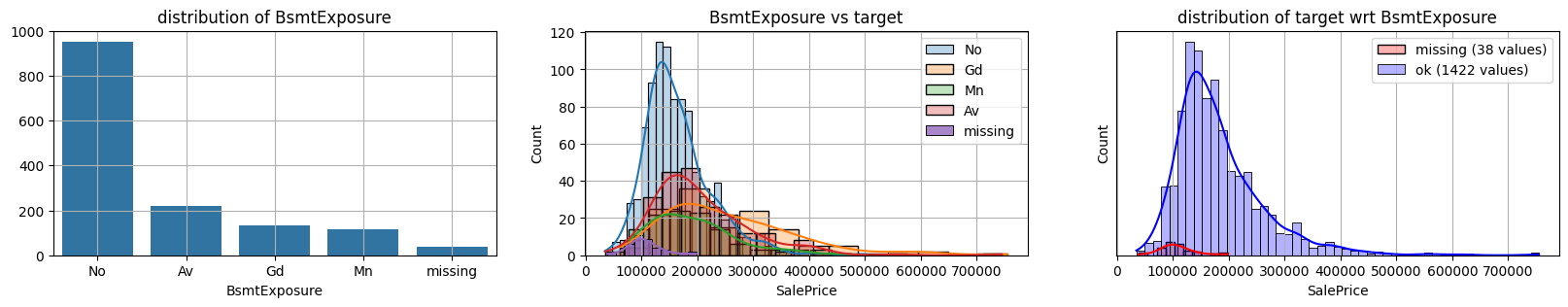
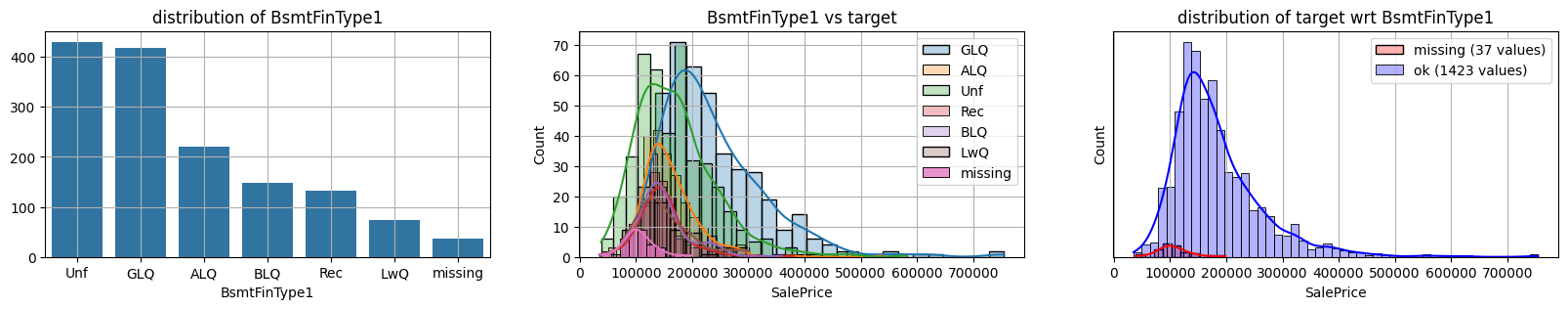
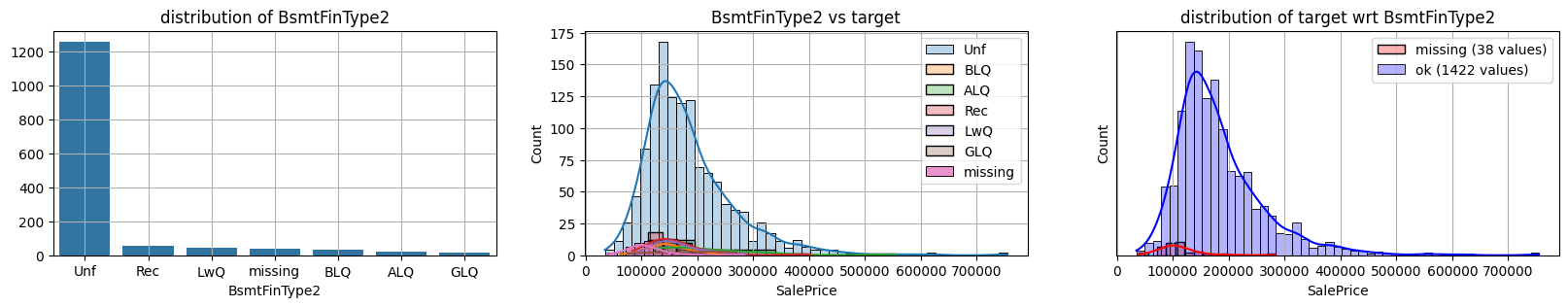
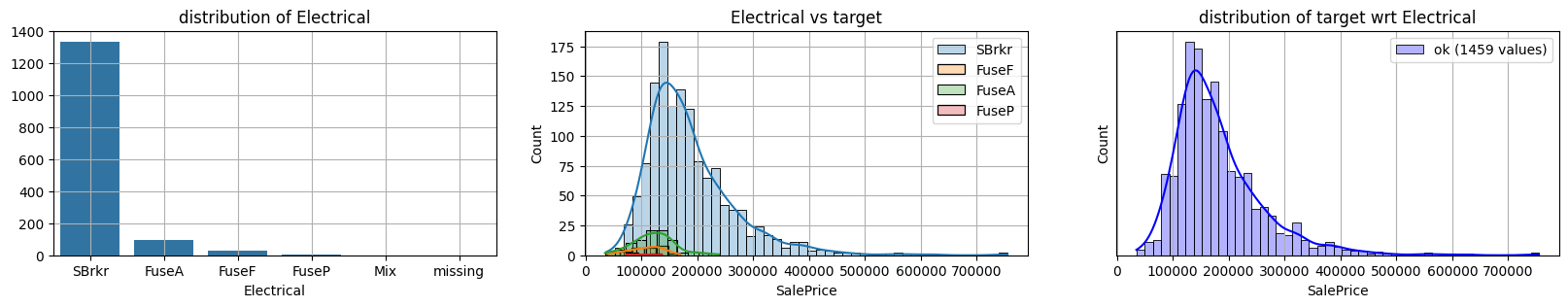
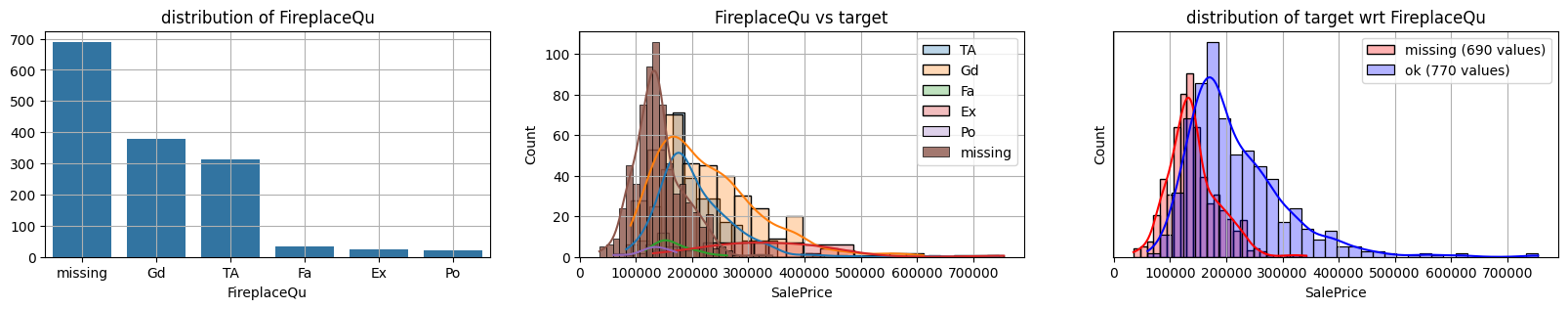
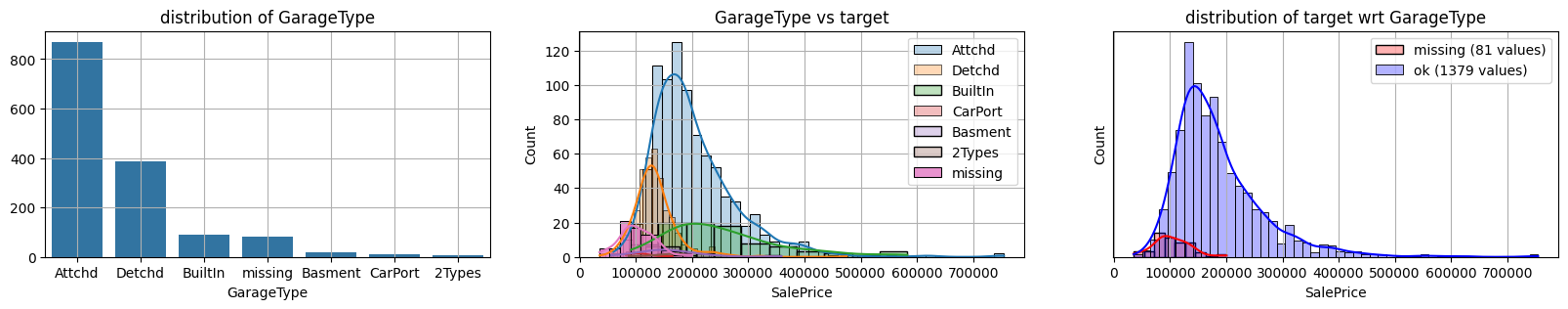
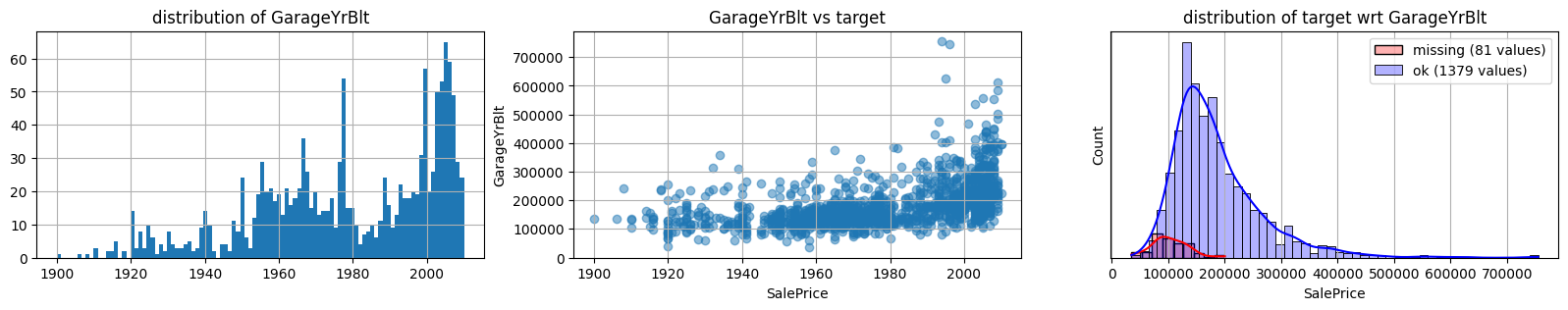
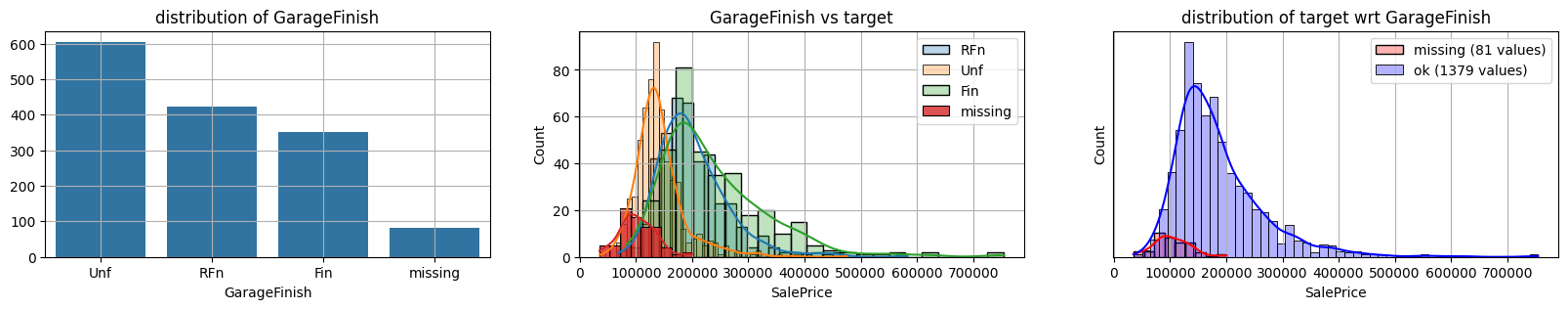
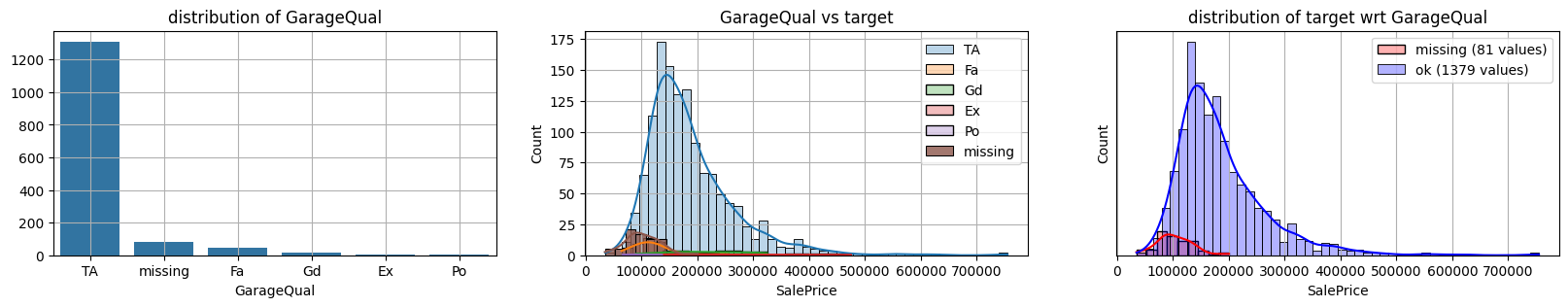
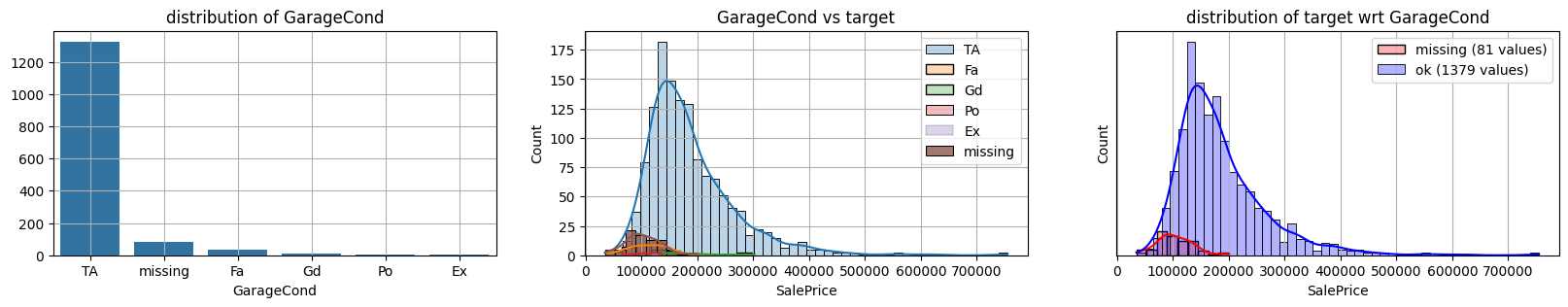
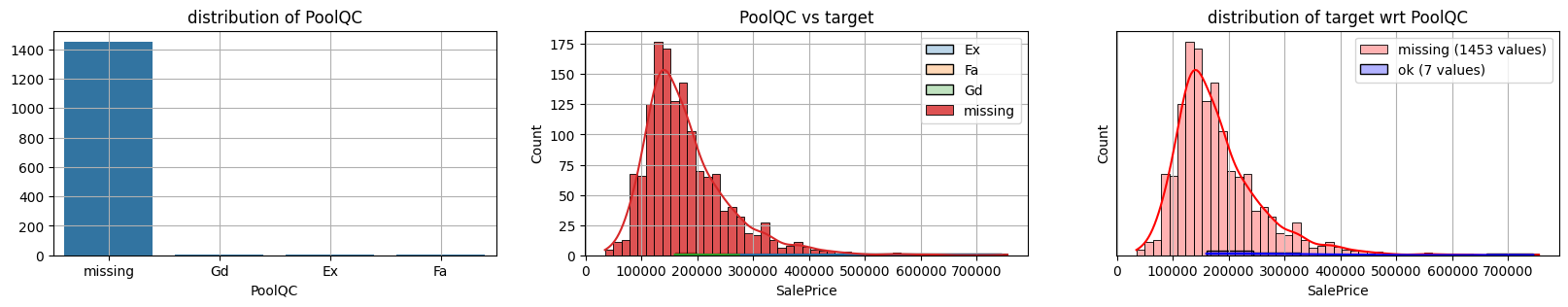
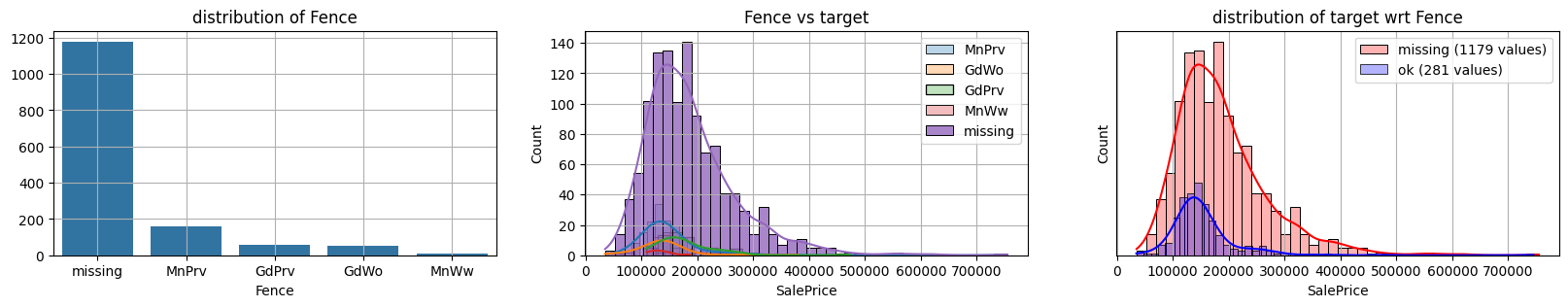
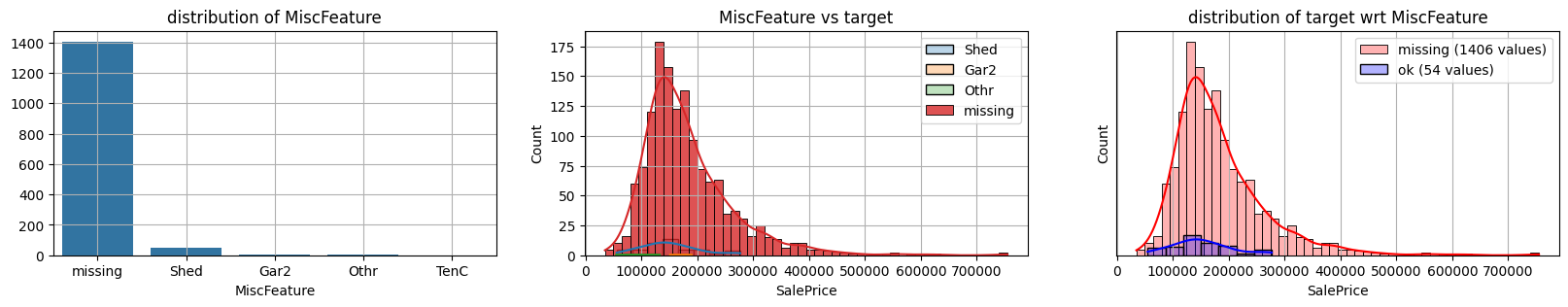
common sense#
too many missing data in Alley. Information might only help non-missing items with little impact on
missing data in Bsmt* seem all the same
missing data in Garage* sell all the same
For continuous variables#
Three substitution techniques#
by a fixed value (zero)
by a fixed value (the mean)
sampling from an equivalent normal (same mean and std)
First we create the different datasets:
dn: original data only with numerical attributesdl0: substituting missing values with zerodlm: substituting missing values with the meandlr: substituting missing values with an equivalent normal (same mean and stdev)
def xdistplot(k, title="", xlim=None):
vals = k
sns.histplot(k, alpha= .8);
m,s = np.mean(vals), np.std(vals)
plt.axvline(m, color="black", lw=2, alpha=.5)
plt.axvline(m+s, color="red", lw=2, alpha=.5)
plt.axvline(m-s, color="red", lw=2, alpha=.5)
x = np.linspace(np.min(vals), np.max(vals), 100)
plt.title(title)
plt.grid();
if xlim is not None:
plt.xlim(xlim)
def subs_policies(d, col):
mcol = "%s_missing"%col
dn = d.T.dropna().T
dn = dn[[i for i in dn.columns if d[i].dtype!=object]]
print (dn.shape)
na_idxs = np.argwhere(d[col].isna().values)[:,0]
dl0 = dn.copy()
dlm = dn.copy()
dlr = dn.copy()
dl0[mcol] = d[col].fillna(0)
dlm[mcol] = d[col].fillna( d[col].mean())
k = d[col].copy()
k[k.isna()] = np.random.normal(loc=np.mean(k), scale=np.std(k), size=np.sum(k.isna()))
dlr[mcol] = k
f0 = lambda: xdistplot(d[col].dropna(), "original", [0,150])
f1 = lambda: xdistplot(dl0[mcol], "subs by zero", [0,150])
f2 = lambda: xdistplot(dlm[mcol], "subs by mean", [0,150])
f3 = lambda: xdistplot(dlr[mcol], "subs by equivalent normal", [0,150])
mlutils.figures_grid(4,1, [f0, f1, f2, f3], figsize=(20,3))
return dn, dl0, dlm, dlr, na_idxs
## KEEPOUTPUT
dn, dl0, dlm, dlr, na_idxs = subs_policies(d, "LotFrontage")
(1460, 34)
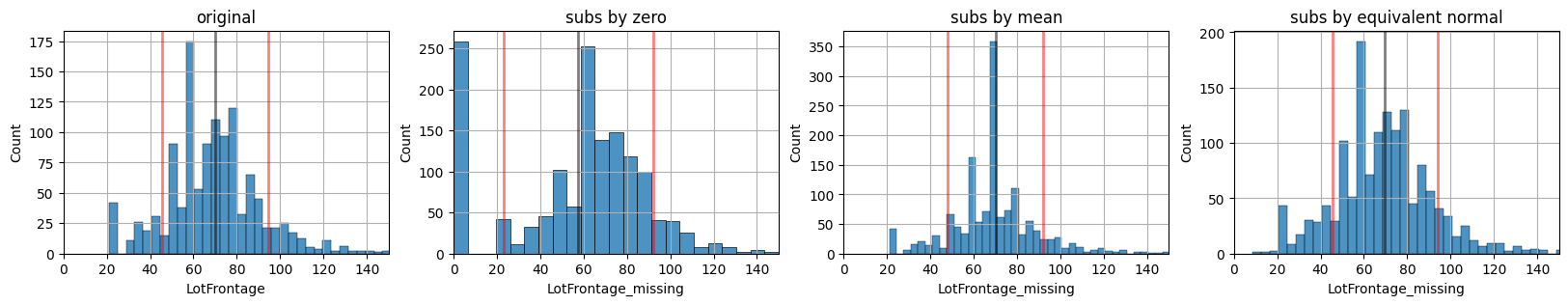
Validation workflow for repairing missing values on LotFrontage#
Which policy for repairing missing data is best?
Short answer: we do not know \(\rightarrow\) we must seek evidence
We will now integrate them in an ML workflow, creating predictive models and seeking for evidence if models improve or not when using different policies for repairing missing data.
We train a lot of models (resampling training data) with each dataset and then run a classical hypothesis test on model performance:
\(e_1\): control group, models trained without LotFrontage
\(e_2\): population group, models trained with LotFrontage with fillna=0
Our null hypothesis (there is no effect in using the new variable):
Our test hypothesis (including fillna=0 improves models):
from sklearn.model_selection import cross_val_score, ShuffleSplit
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error, make_scorer
from scipy.stats import ttest_ind
def getXY (dn):
xcols = [i for i in dn.columns if i!="SalePrice"]
X = dn[xcols].values.astype(float)
y = dn.SalePrice.values.astype(float)
return X,y,xcols
def experiment(dn, estimator, n_models=20, test_size=.3):
X,y,_ = getXY(dn)
r = cross_val_score(estimator, X, y, cv=ShuffleSplit(n_models, test_size=test_size),
scoring=make_scorer(mean_absolute_error))
return r
def HTest(ref_dataset, h_datasets, n_models=30, experiment=experiment, **kwargs):
estimator = RandomForestRegressor(n_estimators=20)
re = [experiment(i, estimator, n_models=n_models, **kwargs) for i in pbar([ref_dataset]+h_datasets)]
for r in re[1:]:
print (ttest_ind(re[0],r))
## KEEPOUTPUT
dl0.head()
| MSSubClass | LotArea | OverallQual | OverallCond | YearBuilt | YearRemodAdd | BsmtFinSF1 | BsmtFinSF2 | BsmtUnfSF | TotalBsmtSF | ... | OpenPorchSF | EnclosedPorch | 3SsnPorch | ScreenPorch | PoolArea | MiscVal | MoSold | YrSold | SalePrice | LotFrontage_missing | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Id | |||||||||||||||||||||
| 1 | 60 | 8450 | 7 | 5 | 2003 | 2003 | 706 | 0 | 150 | 856 | ... | 61 | 0 | 0 | 0 | 0 | 0 | 2 | 2008 | 208500 | 65.0 |
| 2 | 20 | 9600 | 6 | 8 | 1976 | 1976 | 978 | 0 | 284 | 1262 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 2007 | 181500 | 80.0 |
| 3 | 60 | 11250 | 7 | 5 | 2001 | 2002 | 486 | 0 | 434 | 920 | ... | 42 | 0 | 0 | 0 | 0 | 0 | 9 | 2008 | 223500 | 68.0 |
| 4 | 70 | 9550 | 7 | 5 | 1915 | 1970 | 216 | 0 | 540 | 756 | ... | 35 | 272 | 0 | 0 | 0 | 0 | 2 | 2006 | 140000 | 60.0 |
| 5 | 60 | 14260 | 8 | 5 | 2000 | 2000 | 655 | 0 | 490 | 1145 | ... | 84 | 0 | 0 | 0 | 0 | 0 | 12 | 2008 | 250000 | 84.0 |
5 rows × 35 columns
## KEEPOUTPUT
HTest(dn, [dl0, dlm, dlr], n_models=50)
100% (4 of 4) |##########################| Elapsed Time: 0:00:49 Time: 0:00:49
TtestResult(statistic=-1.3541252006646298, pvalue=0.1788107411367549, df=98.0)
TtestResult(statistic=-0.6937964889158504, pvalue=0.48945111677260755, df=98.0)
TtestResult(statistic=-0.5677251272549229, pvalue=0.5715199997392869, df=98.0)
## KEEPOUTPUT
HTest(dn, [dl0, dlm, dlr], n_models=50)
100% (4 of 4) |##########################| Elapsed Time: 0:00:49 Time: 0:00:49
TtestResult(statistic=-0.37511561454083137, pvalue=0.7083850959273256, df=98.0)
TtestResult(statistic=-0.3321948468400306, pvalue=0.7404516048445651, df=98.0)
TtestResult(statistic=0.4780530200131818, pvalue=0.6336771343561804, df=98.0)
No \(p\) value is really significative so the different substitution policies do no help improve overall in this simplified setting. More over, repeated experiments show to evidence of any approach better than others (always better \(p\) value)
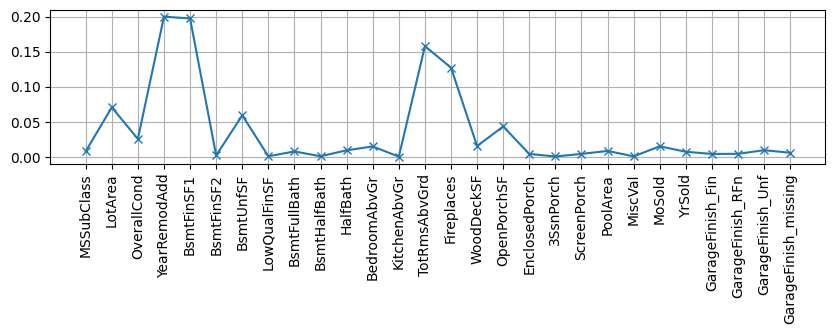
Some models provide us with a measure of feature importance. Observe the behaviour of the most important variables in the scatter matrix plot of previous notebook.
## KEEPOUTPUT
X, y, xcols = getXY(dlr)
rf = RandomForestRegressor(n_estimators=20)
rf.fit(X,y);
plt.figure(figsize=(10,2)); plt.grid()
plt.plot(rf.feature_importances_, label="est1", marker="x")
plt.xticks(range(len(xcols)), xcols, rotation="vertical");
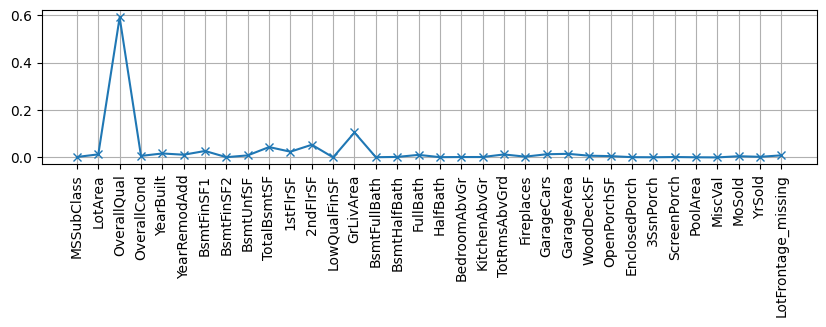
so somehow it is understandable that providing values for this missing variable does not have so much impact.
We could try to see ONLY how records with THIS missing data behave. Observe that na_idxs marks which rows contain na values so that they are used to measure validation performance.
There is still no clear evidence that any filling strategy for missing values is better than other.
def na_cross_val_score(estimator, X, y, cv, scoring, val_idxs):
r = []
for tr_idxs, ts_idxs in cv.split(X):
tr_idxs, ts_idxs = np.r_[tr_idxs], np.r_[ts_idxs]
rf.fit(X[tr_idxs], y[tr_idxs])
valts_idxs = np.r_[[i for i in ts_idxs if i in val_idxs]]
r.append(scoring(rf, X[valts_idxs], y[valts_idxs]))
return r
def na_experiment(dn, estimator, na_idxs, n_models=20, test_size=.3):
X,y,_ = getXY(dn)
r = na_cross_val_score(estimator, X, y, cv=ShuffleSplit(n_models, test_size=test_size),
scoring=make_scorer(mean_absolute_error), val_idxs=na_idxs)
return r
## KEEPOUTPUT
HTest(dn, [dl0, dlm, dlr], experiment=na_experiment, na_idxs=na_idxs, n_models=50)
100% (4 of 4) |##########################| Elapsed Time: 0:00:49 Time: 0:00:49
TtestResult(statistic=-2.5660466465640033, pvalue=0.011800717881861248, df=98.0)
TtestResult(statistic=-2.053620290722926, pvalue=0.042675908653560114, df=98.0)
TtestResult(statistic=-1.5568314871351756, pvalue=0.12273414960890429, df=98.0)
HTest(dn, [dl0, dlm, dlr], experiment=na_experiment, na_idxs=na_idxs, n_models=50)
100% (4 of 4) |##########################| Elapsed Time: 0:00:49 Time: 0:00:49
TtestResult(statistic=-1.4547828358181458, pvalue=0.14892498057497006, df=98.0)
TtestResult(statistic=-0.5523471899998421, pvalue=0.5819680699474099, df=98.0)
TtestResult(statistic=-1.4511464404586971, pvalue=0.14993266003326222, df=98.0)
For categorical features#
we must convert them to numerical
if categories are ordered \(\rightarrow\) convert to positive integer
otherwise \(\rightarrow\) convert to one hot
we must decide on missing values:
remove row or column
assign an existing value
assign a new value
## KEEPOUTPUT
col = "GarageFinish"
print ("missing", sum(d[col].isna()))
d[col].value_counts()
missing 81
GarageFinish
Unf 605
RFn 422
Fin 352
Name: count, dtype: int64
def to_onehot(x):
values = np.unique(x)
r = np.r_[[np.argwhere(i==values)[0][0] for i in x]]
return np.eye(len(values))[r].astype(int)
def replace_column_with_onehot(d, col):
assert sum(d[col].isna())==0, "column must have no NaN values"
values = np.unique(d[col]
)
k = to_onehot(d[col].values)
r = pd.DataFrame(k, columns=["%s_%s"%(col, values[i]) for i in range(k.shape[1])], index=d.index).join(d)
del(r[col])
return r
observe onehot encoding
## KEEPOUTPUT
d[[col]].head(10)
| GarageFinish | |
|---|---|
| Id | |
| 1 | RFn |
| 2 | RFn |
| 3 | RFn |
| 4 | Unf |
| 5 | RFn |
| 6 | Unf |
| 7 | RFn |
| 8 | RFn |
| 9 | Unf |
| 10 | RFn |
## KEEPOUTPUT
replace_column_with_onehot(d[[col]].dropna().copy(), col).head(10)
| GarageFinish_Fin | GarageFinish_RFn | GarageFinish_Unf | |
|---|---|---|---|
| Id | |||
| 1 | 0 | 1 | 0 |
| 2 | 0 | 1 | 0 |
| 3 | 0 | 1 | 0 |
| 4 | 0 | 0 | 1 |
| 5 | 0 | 1 | 0 |
| 6 | 0 | 0 | 1 |
| 7 | 0 | 1 | 0 |
| 8 | 0 | 1 | 0 |
| 9 | 0 | 0 | 1 |
| 10 | 0 | 1 | 0 |
we now create a onehot encoding for each case:
create a separate value for missing data
set missing data to an existing category. In this case we will set it to the closest category distribution wrt the target variable according the plots above
## KEEPOUTPUT
rm1 = replace_column_with_onehot(d[[col]].fillna("missing").copy(), col)
rm2 = replace_column_with_onehot(d[[col]].fillna("Unf").copy(), col)
## KEEPOUTPUT
rm1.head()
| GarageFinish_Fin | GarageFinish_RFn | GarageFinish_Unf | GarageFinish_missing | |
|---|---|---|---|---|
| Id | ||||
| 1 | 0 | 1 | 0 | 0 |
| 2 | 0 | 1 | 0 | 0 |
| 3 | 0 | 1 | 0 | 0 |
| 4 | 0 | 0 | 1 | 0 |
| 5 | 0 | 1 | 0 | 0 |
## KEEPOUTPUT
rm2.head()
| GarageFinish_Fin | GarageFinish_RFn | GarageFinish_Unf | |
|---|---|---|---|
| Id | |||
| 1 | 0 | 1 | 0 |
| 2 | 0 | 1 | 0 |
| 3 | 0 | 1 | 0 |
| 4 | 0 | 0 | 1 |
| 5 | 0 | 1 | 0 |
## KEEPOUTPUT
dm1 = dn.join(rm1)
dm2 = dn.join(rm2)
dm1.shape, dm2.shape
((1460, 38), (1460, 37))
again, do hypothesis testing to seek for evidence for improvements for any of the methods
## KEEPOUTPUT
HTest(dn, [dm1, dm2], experiment=experiment, n_models=50)
100% (3 of 3) |##########################| Elapsed Time: 0:00:38 Time: 0:00:38
TtestResult(statistic=-0.7343576845947623, pvalue=0.4644842464733756, df=98.0)
TtestResult(statistic=-0.35926068145867207, pvalue=0.7201730079370724, df=98.0)
## KEEPOUTPUT
na_idxs = np.argwhere(d[col].isna().values)[:,0]
HTest(dn, [dm1, dm2], experiment=na_experiment, na_idxs=na_idxs, n_models=50)
100% (3 of 3) |##########################| Elapsed Time: 0:00:37 Time: 0:00:37
TtestResult(statistic=2.0301224227097934, pvalue=0.045054029061182464, df=98.0)
TtestResult(statistic=1.1337018450245944, pvalue=0.25968538695450255, df=98.0)
Again, no \(p\) value is sufficiently significative to provide evidence for improved classification alone. However, approach number 2 (substituting missing data with Unf) does seem to consistently get better \(p\) value and, thus, more chance to improve performance when combined with other data cleaning choices.
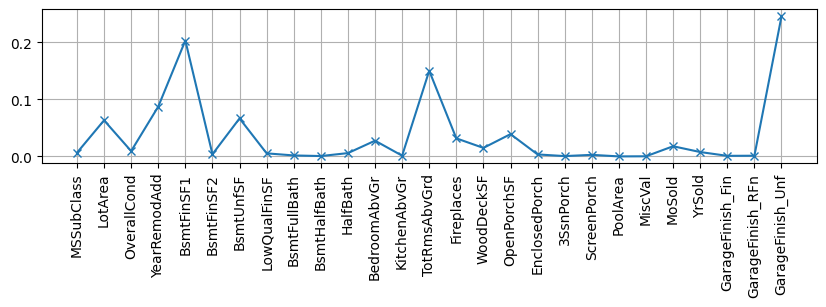
Observe also how the missing category dilutes the importance of Unf after other variables are taken out.
## KEEPOUTPUT
dk = dm2.copy()
dk = dk[[i for i in dm2.columns if not i in ["OverallQual", "GarageCars", "GrLivArea",
"GarageArea", "YearBuilt", "TotalBsmtSF",
"1stFlrSF", "2ndFlrSF", "FullBath"]]]
X, y, xcols = getXY(dk)
rf = RandomForestRegressor(n_estimators=10)
rf.fit(X,y);
plt.figure(figsize=(10,2)); plt.grid()
plt.plot(rf.feature_importances_, label="est1", marker="x")
plt.xticks(range(len(xcols)), xcols, rotation="vertical");
## KEEPOUTPUT
dk = dm1.copy()
dk = dk[[i for i in dm1.columns if not i in ["OverallQual", "GarageCars", "GrLivArea",
"GarageArea", "YearBuilt", "TotalBsmtSF",
"1stFlrSF", "2ndFlrSF", "FullBath"]]]
X, y, xcols = getXY(dk)
rf = RandomForestRegressor(n_estimators=10)
rf.fit(X,y);
plt.figure(figsize=(10,2)); plt.grid()
plt.plot(rf.feature_importances_, label="est1", marker="x")
plt.xticks(range(len(xcols)), xcols, rotation="vertical");