06.03 - PCA, NMF IN PRACTICE#
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
Reducción de dimensionalidad para tareas de clasificación#
mnist = pd.read_csv("local/data/mnist1.5k.csv.gz", compression="gzip", header=None).values
d=mnist[:,1:785]
c=mnist[:,0]
print ("dimension de las imagenes y las clases", d.shape, c.shape)
dimension de las imagenes y las clases (1500, 784) (1500,)
plt.imshow(d[9].reshape(28,28), cmap=plt.cm.gray)
<matplotlib.image.AxesImage at 0x7fa29c7058e0>
perm = np.random.permutation(range(d.shape[0]))[0:50]
random_imgs = d[perm]
random_labels = c[perm]
fig = plt.figure(figsize=(10,6))
for i in range(random_imgs.shape[0]):
ax=fig.add_subplot(5,10,i+1)
plt.imshow(random_imgs[i].reshape(28,28), interpolation="nearest", cmap = plt.cm.Greys_r)
ax.set_title(int(random_labels[i]))
ax.set_xticklabels([])
ax.set_yticklabels([])
Principal Component Analysis#
from sklearn.decomposition import PCA
mnist = pd.read_csv("local/data/mnist1.5k.csv.gz", compression="gzip", header=None).values
X=mnist[:,1:785]
y=mnist[:,0]
pca = PCA(n_components=10)
Xp = pca.fit_transform(X)
X.shape, Xp.shape
((1500, 784), (1500, 10))
for i in np.unique(y):
print (i, np.sum(y==i))
0 150
1 157
2 186
3 125
4 151
5 138
6 152
7 154
8 141
9 146
from sklearn.model_selection import train_test_split
Xtr, Xts, ytr, yts = train_test_split(X,y,test_size=.3)
Xtr.shape, Xts.shape, ytr.shape, yts.shape
((1050, 784), (450, 784), (1050,), (450,))
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
dt = GaussianNB()
dt.fit(Xtr, ytr)
dt.score(Xtr, ytr), dt.score(Xts, yts)
(0.72, 0.6177777777777778)
cs = range(10,200,5)
dtr, dts = [], []
for n_components in cs:
print (".", end="")
pca = PCA(n_components=n_components)
pca.fit(Xtr)
Xt_tr = pca.transform(Xtr)
Xt_ts = pca.transform(Xts)
dt.fit(Xt_tr,ytr)
ypreds_tr = dt.predict(Xt_tr)
ypreds_ts = dt.predict(Xt_ts)
ypreds_tr.shape, ypreds_ts.shape
dtr.append(np.mean(ytr==ypreds_tr))
dts.append(np.mean(yts==ypreds_ts))
......................................
len(dtr), len(dts)
(38, 38)
plt.plot(cs, dtr, label="train")
plt.plot(cs, dts, label="test")
plt.xlabel("n components")
plt.ylabel("% acierto")
plt.legend()
<matplotlib.legend.Legend at 0x7f5316c9c790>
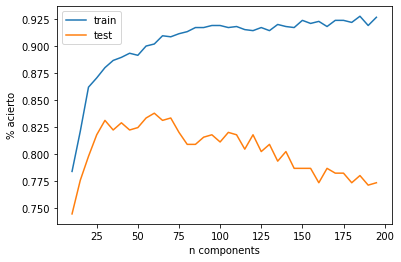
best_cs = cs[np.argmax(dts)]
best_cs
60
clasificación en el nuevo espacio de representación#
pca = PCA(n_components=best_cs)
pca.fit(Xtr)
Xt_tr = pca.transform(Xtr)
Xt_ts = pca.transform(Xts)
dt.fit(Xt_tr,ytr)
ypreds_tr = dt.predict(Xt_tr)
ypreds_ts = dt.predict(Xt_ts)
ypreds_tr.shape, ypreds_ts.shape
np.mean(ytr==ypreds_tr),np.mean(yts==ypreds_ts)
(0.9047619047619048, 0.8355555555555556)
pipelines#
debemos de tener cuidado cuando usamos transformaciones en clasificación, ya que tenemos que ajustarlas (de manera no supervisada) sólo con los datos de entrenamiento
from sklearn.pipeline import Pipeline
estimator = Pipeline((("pca", PCA(n_components=best_cs)), ("naive", dt)))
estimator.fit(Xtr, ytr)
estimator.score(Xtr, ytr), estimator.score(Xts, yts)
(0.9057142857142857, 0.8333333333333334)
from sklearn.model_selection import cross_val_score
pip = Pipeline([("PCA", PCA(n_components=best_cs)), ("gaussian", GaussianNB())])
scores = cross_val_score(pip, X,y, cv=5 )
print ("%.2f +/- %.4f"%(np.mean(scores), np.std(scores)))
0.84 +/- 0.0168
obtenemos los componentes principales#
cols=20
plt.figure(figsize=(15,3))
for i in range(len(pca.components_)):
plt.subplot(np.ceil(len(pca.components_)/15.),15,i+1)
plt.imshow((pca.components_[i].reshape(28,28)), cmap = plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

verificamos la reconstrucción con los componentes principales#
pca = PCA(n_components=best_cs)
pca.fit(Xtr)
Xp = pca.transform(X)
plt.figure(figsize=(10,6))
for i in range(6):
plt.subplot(3,6,i+1)
k = np.random.randint(len(X))
plt.imshow((np.sum((pca.components_*Xp[k].reshape(-1,1)), axis=0)).reshape(28,28), cmap=plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])
plt.subplot(3,6,6+i+1)
plt.imshow(X[k].reshape(28,28), cmap=plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

observa la nueva representación de la primera imagen#
X[0]
array([ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 188, 255, 94, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 191, 250, 253, 93, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 123, 248, 253, 167, 10, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 80, 247, 253, 208, 13, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 29, 207, 253, 235, 77, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 54, 209, 253, 253, 88, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 93, 254, 253, 238, 170,
17, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 23, 210, 254, 253,
159, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 16, 209, 253,
254, 240, 81, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 27,
253, 253, 254, 13, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
20, 206, 254, 254, 198, 7, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 168, 253, 253, 196, 7, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 20, 203, 253, 248, 76, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 22, 188, 253, 245, 93, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 103, 253, 253, 191, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 89, 240, 253, 195, 25, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 15, 220, 253, 253, 80,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 94, 253, 253,
253, 94, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 89,
251, 253, 250, 131, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 214, 218, 95, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0])
Xp[0]
array([-602.96544279, 709.91417575, 48.46393261, -94.3578192 ,
-247.5969329 , -429.06089585, -197.60517121, 531.22635527,
439.83674877, -143.65598588, -350.95470187, 172.60373135,
-172.73696352, -281.72027853, -126.34179342, 161.73658414,
-81.78527347, -413.79147679, 47.59742414, -237.42715084,
223.18870607, -258.81025144, 43.07849287, -144.98480884,
-56.84875879, 15.95569482, -37.29026752, -169.61029386,
290.04985091, -18.09769242, -156.47270457, 115.19250934,
-92.38887583, 97.42047216, -165.2477172 , 195.55535288,
202.84586027, 19.77436206, -103.13657262, -196.06966884,
124.32311744, 31.23761748, -38.67664165, 35.31148678,
70.87604485, 186.20659945, -128.96485438, 66.94065907,
-149.08882182, -46.65103309, -4.09849795, 111.30817383,
-52.76418032, -40.0893952 , -70.94692439, -105.78083452,
-23.79724802, 152.12095565, 59.00594154, 7.56383813])
which correspond to the same components above for PCA.
Non negative matrix factorization#
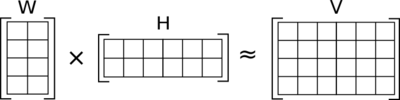
Descomponemos una matriz \(V \in \mathbb{R}_+^{m\times n}\) en el producto \(W \times H\), con \(W \in \mathbb{R}_+^{m\times r}\) y \(H \in \mathbb{R}_+^{r\times n}\) con la restricción de que todo sea positivo (\(\in \mathbb{R}_+\)), de forma que:
Las filas de \(H\) son los componentes base, y se soluciona planteándolo como un problema de optimización matemática con restricciones.
from IPython.display import Image
Image(filename='local/imgs/nmf.png')
obtenemos la descomposición#
from sklearn.decomposition import NMF
X=mnist[:,1:785]; y=mnist[:,0]
nmf = NMF(n_components=15, init="random")
Xn = nmf.fit_transform(X)
cols=20
plt.figure(figsize=(15,3))
for i in range(len(nmf.components_)):
plt.subplot(len(nmf.components_)/15,15,i+1)
plt.imshow(np.abs(nmf.components_[i].reshape(28,28)), cmap = plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

Xn[0,:]
array([0. , 0. , 4.95514551, 0. , 0. ,
6.08083653, 1.91867923, 0.09777909, 0. , 0. ,
0. , 8.73915575, 0. , 3.50861945, 0. ])
verfiicamos la reconstrucción#
plt.figure(figsize=(10,6))
for i in range(6):
plt.subplot(3,6,i+1)
k = np.random.randint(len(X))
plt.imshow(np.abs(np.sum((nmf.components_*Xn[k].reshape(-1,1)), axis=0)).reshape(28,28), cmap=plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])
plt.subplot(3,6,6+i+1)
plt.imshow(X[k].reshape(28,28), cmap=plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

clasificamos en el nuevo espacio de representación#
print (np.mean(cross_val_score(GaussianNB(), X,y, cv=5 )))
print (np.mean(cross_val_score(GaussianNB(), Xn,y, cv=5 )))
0.5953333333333334
0.7733333333333333
la primera imagen en el nuevo espacio de representación#
observa que todos los componentes son positivos
plt.imshow(X[0].reshape(28,28))
<matplotlib.image.AxesImage at 0x7f5315e32190>

Xn[0]
array([0. , 0. , 4.95514551, 0. , 0. ,
6.08083653, 1.91867923, 0.09777909, 0. , 0. ,
0. , 8.73915575, 0. , 3.50861945, 0. ])
cols=20
plt.figure(figsize=(15,3))
for i in range(len(nmf.components_)):
plt.subplot(len(nmf.components_)/15,15,i+1)
plt.imshow(np.abs(nmf.components_[i].reshape(28,28)), cmap = plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

NMF para el reconocimiento de rostros#
import numpy as np
faces = np.load("local/data/faces.npy")
plt.figure(figsize=(15,2))
for i in range(30):
plt.subplot(2,15,i+1)
plt.imshow(faces[np.random.randint(len(faces))].reshape(19,19), cmap=plt.cm.Greys_r)
plt.xticks([]); plt.yticks([])

nmf = NMF(n_components=30, init="random")
faces_n = nmf.fit_transform(faces)
cols=20
plt.figure(figsize=(15,2))
for i in range(len(nmf.components_)):
plt.subplot(np.ceil(len(nmf.components_)/15.),15,i+1)
plt.imshow(np.abs(nmf.components_[i].reshape(19,19)), cmap = plt.cm.Greys)
plt.xticks([]); plt.yticks([])

forzamos dispersión en los componentes, y extendemos el problema de optimización con la norma \(L_1\) en los componentes base.
también podríamos forzar dispersión en la nueva representación $\(\begin{split} argmin_{W,H}\;& ||V-W\times H|| + ||W||^2_1\\ s.t.&\;W,H \in \mathbb{R}_+ \end{split}\)$
nmf = NMF(n_components=30, init="nndsvd", alpha=1000, l1_ratio=1)
faces_n = nmf.fit_transform(faces)
cols=20
plt.figure(figsize=(15,2))
print (np.sum(nmf.components_))
for i in range(len(nmf.components_)):
plt.subplot(np.ceil(len(nmf.components_)/15.),15,i+1)
plt.imshow(np.abs(nmf.components_[i].reshape(19,19)), cmap = plt.cm.Greys)
plt.xticks([]); plt.yticks([])
14650.57060382788


