04.04 - COMPARING SAMPLES#
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
endpoint https://m5knaekxo6.execute-api.us-west-2.amazonaws.com/dev-v0001/rlxmooc
from scipy import stats
import numpy as np
import matplotlib.pyplot as plt
from progressbar import progressbar as pbar
%matplotlib inline
Comparing populations. A two-sided test#
We have some exam scores for two groups of 10 students each
STEP 1: Define the model you want to challenge (the NULL Hypothesis \(H_0\))#
We define the \(H_0\) as a model where there is no statistical difference between the two groups of students.
STEP 2: Define your REAL WORLD sample and your test statistic#
SAMPLE: we sample two groups of 10 students exam scores each. We KNOW there is a difference (
mean_diff) because we create the use case in such way. We want to see if the test detects it.Can it detect it with larger/smaller group sizes?
Can it detect it with larger/smaller mean differences?
real_world_sample_size = 10
mu, sigma = 100, 10
def generate_AB(mu=mu, sigma=sigma, real_world_sample_size=real_world_sample_size, mean_diff=5):
A = np.random.normal(loc=mu, scale=sigma, size=real_world_sample_size)
B = np.random.normal(loc=mu+mean_diff, scale=sigma, size=real_world_sample_size)
return A, B
A,B = generate_AB()
print ("group A [", " ".join(["%6.2f"%i for i in A]), "] mean %6.2f"%np.mean(A))
print ("group B [", " ".join(["%6.2f"%i for i in B]), "] mean %6.2f"%np.mean(B))
group A [ 96.08 101.76 89.45 88.81 106.66 105.94 95.99 108.45 94.67 114.32 ] mean 100.21
group B [ 129.23 127.05 115.34 105.17 117.75 121.96 103.69 120.42 118.77 89.56 ] mean 114.89
if we look at these two sets of scores, how sure can we be that their respective populations are different (different \(\mu\))?
Define a test statistic to measure the mean difference of the samples#
where:
\(\bar{A}\), \(\bar{B}\) is the mean of the two real world samples
\(S_A\), \(S_B\) is the standard deviation of the two real world samples
\(N_A\), \(N_B\) is the size (number of elementso) of the two real world samples
The denominator factor is included so that we can compare the means with different standard deviations.
STEP 3: We simulate \(H_0\), and understand its distribution of \(\text{ttest}\)#
individual student scores NEED NOT come from a normal disitribution. Regardless their origin, the distribution of the test statistic will be normal.
ttest = lambda a,b: (np.mean(a)-np.mean(b))/np.sqrt(np.std(a)**2/len(a)+np.std(b)**2/len(b))
n = 3000
a = np.r_[[stats.norm(loc=mu, scale=sigma).rvs(real_world_sample_size) for _ in pbar(range(n))]]
b = np.r_[[stats.norm(loc=mu, scale=sigma).rvs(real_world_sample_size) for _ in pbar(range(n))]]
t = np.r_[[ttest(i,j) for i,j in zip(a,b)]]
100% (3000 of 3000) |####################| Elapsed Time: 0:00:01 Time: 0:00:01
100% (3000 of 3000) |####################| Elapsed Time: 0:00:01 Time: 0:00:01
# A,B = generate_AB()
plt.hist( t, density=True, alpha=.5, bins=30);
plt.axvline(np.percentile(t,2.5), color="black", label="%5 two sided interval\n(2.5% - 97.5%) ")
plt.axvline(np.percentile(t,97.5), color="black")
plt.grid(); plt.legend();
plt.xlabel("difference of means"); plt.ylabel("probability")
plt.show()
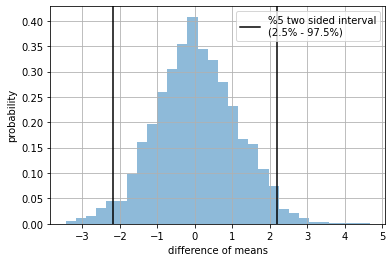
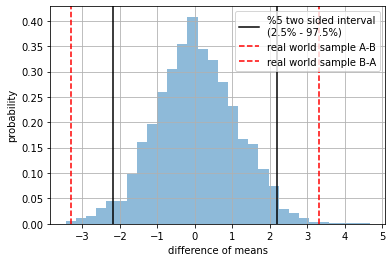
STEP 5: How rare is our real world sample w.r.t. \(H_0\)?#
we measure how much probability mass is left outside the red dashed lines (our real world sample)
small \(p_{value}\) means our real world sample is rare
large \(p_{value}\) means our real world sample is quite common (expected)
For a confidence interval \(\alpha=0.05\), if \(p_{value}<\alpha\) we reject \(H_0\) and consider \(A\) and \(B\) actually come from different populations.
observe we display \(\text{ttest}(A,B)\) and \(\text{ttest}(B,A)\) (red dashed lines) since we want to check if they are different in any direction
print ("group A [", " ".join(["%6.2f"%i for i in A]), "] mean %6.2f"%np.mean(A))
print ("group B [", " ".join(["%6.2f"%i for i in B]), "] mean %6.2f"%np.mean(B))
plt.hist( t, density=True, alpha=.5, bins=30);
plt.axvline(np.percentile(t,2.5), color="black", label="%5 two sided interval\n(2.5% - 97.5%) ")
plt.axvline(np.percentile(t,97.5), color="black")
plt.axvline(ttest(A,B), color="red", ls="--", label="real world sample A-B")
plt.axvline(ttest(B,A), color="red", ls="--", label="real world sample B-A")
plt.grid(); plt.legend();
plt.xlabel("difference of means"); plt.ylabel("probability")
plt.show()
group A [ 96.08 101.76 89.45 88.81 106.66 105.94 95.99 108.45 94.67 114.32 ] mean 100.21
group B [ 129.23 127.05 115.34 105.17 117.75 121.96 103.69 120.42 118.77 89.56 ] mean 114.89
#A,B = generate_AB()
k = np.mean( ttest(A,B)<t)
k = (k if k<0.5 else 1-k)*2
print ("empirical (simulated) p-value %.4f"%k)
empirical (simulated) p-value 0.0013
which corresponds to the value obtained with the analytical formula#
As implemented in scipy.stats.ttest_ind. With larger simulations, the result gets closer to the analytical formula.
stats.ttest_ind(A, B).pvalue
0.005726493310989277
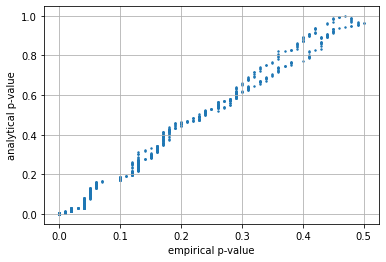
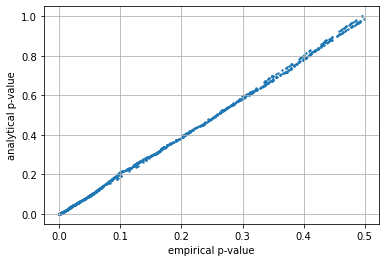
Understanding accuracy of large and small simulations#
observe the relation between the empirical (simulated) and analytical \(p_{value}\) is much tighter (more accurate) when the simulation is larger.
def test_simulations(n, mu=mu, sigma=sigma, real_world_sample_size=real_world_sample_size):
a = np.r_[[stats.norm(loc=mu, scale=sigma).rvs(real_world_sample_size) for _ in pbar(range(n))]]
b = np.r_[[stats.norm(loc=mu, scale=sigma).rvs(real_world_sample_size) for _ in pbar(range(n))]]
t = np.r_[[ttest(A, B) for A,B in zip(a,b)]]
r = []
for _ in range(1000):
A,B = generate_AB(mu=mu, sigma=sigma, real_world_sample_size=real_world_sample_size)
ts = ttest(A,B)
k = np.mean( ts<t)
k = (k if k<0.5 else 1-k)*2
r.append([k, stats.ttest_ind(A, B).pvalue])
r = np.r_[r]
plt.scatter(r[:,0], r[:,1], s=2)
plt.grid(); plt.xlabel("empirical p-value"); plt.ylabel("analytical p-value")
em, an = r[:,0], r[:,1]
print ("p_value mean error simulated/analytic %.3f"%np.mean(np.abs(em-an)))
print ("p_value mean error when p_value<0.05 %.3f"%np.mean(np.abs(em[an<0.05]-an[an<0.05])))
test_simulations(n=100, mu=mu, sigma=sigma)
100% (100 of 100) |######################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (100 of 100) |######################| Elapsed Time: 0:00:00 Time: 0:00:00
p_value mean error simulated/analytic 0.165
p_value mean error when p_value<0.05 0.004
test_simulations(n=1000, mu=mu, sigma=sigma)
100% (1000 of 1000) |####################| Elapsed Time: 0:00:00 Time: 0:00:00
100% (1000 of 1000) |####################| Elapsed Time: 0:00:00 Time: 0:00:00
p_value mean error simulated/analytic 0.172
p_value mean error when p_value<0.05 0.016
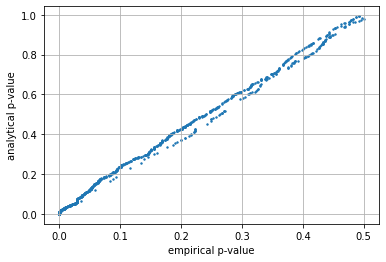
test_simulations(n=6000, mu=mu, sigma=sigma)
100% (6000 of 6000) |####################| Elapsed Time: 0:00:02 Time: 0:00:02
100% (6000 of 6000) |####################| Elapsed Time: 0:00:02 Time: 0:00:02
p_value mean error simulated/analytic 0.168
p_value mean error when p_value<0.05 0.009
For more info:
Sampling distribution of the sample mean Khan Academy
Central limit theorem Wikipedia

