05.02 - MODEL EVALUATION#
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
replicating local resources
endpoint https://m3g87w9l3k.execute-api.us-west-2.amazonaws.com/dev/rlxmooc
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import local.lib.timeseries as ts
from local.lib import calhousing as ch
%matplotlib inline
The cal_housing repository publicly available#
!head local/data/cal_housing_small.data
!wc local/data/cal_housing_small.data
longitude,latitude,housingMedianAge,totalRooms,totalBedrooms,medianHouseValue
-120.58,35.0,37.0,523.0,119.0,106300.0
-118.17,33.98,31.0,1236.0,329.0,155400.0
-122.22,37.81,52.0,1971.0,335.0,273700.0
-117.91,33.66,21.0,1708.0,505.0,193800.0
-121.92,37.24,27.0,1265.0,216.0,281200.0
-117.01,32.71,20.0,3506.0,692.0,129100.0
-116.39,34.15,15.0,5583.0,1149.0,73300.0
-120.67,35.5,15.0,2752.0,546.0,175000.0
-118.18,34.04,36.0,1807.0,630.0,129000.0
501 501 20363 local/data/cal_housing_small.data
d = pd.read_csv("local/data/cal_housing_small.data")
print (d.shape)
d.head()
(500, 6)
| longitude | latitude | housingMedianAge | totalRooms | totalBedrooms | medianHouseValue | |
|---|---|---|---|---|---|---|
| 0 | -120.58 | 35.00 | 37.0 | 523.0 | 119.0 | 106300.0 |
| 1 | -118.17 | 33.98 | 31.0 | 1236.0 | 329.0 | 155400.0 |
| 2 | -122.22 | 37.81 | 52.0 | 1971.0 | 335.0 | 273700.0 |
| 3 | -117.91 | 33.66 | 21.0 | 1708.0 | 505.0 | 193800.0 |
| 4 | -121.92 | 37.24 | 27.0 | 1265.0 | 216.0 | 281200.0 |
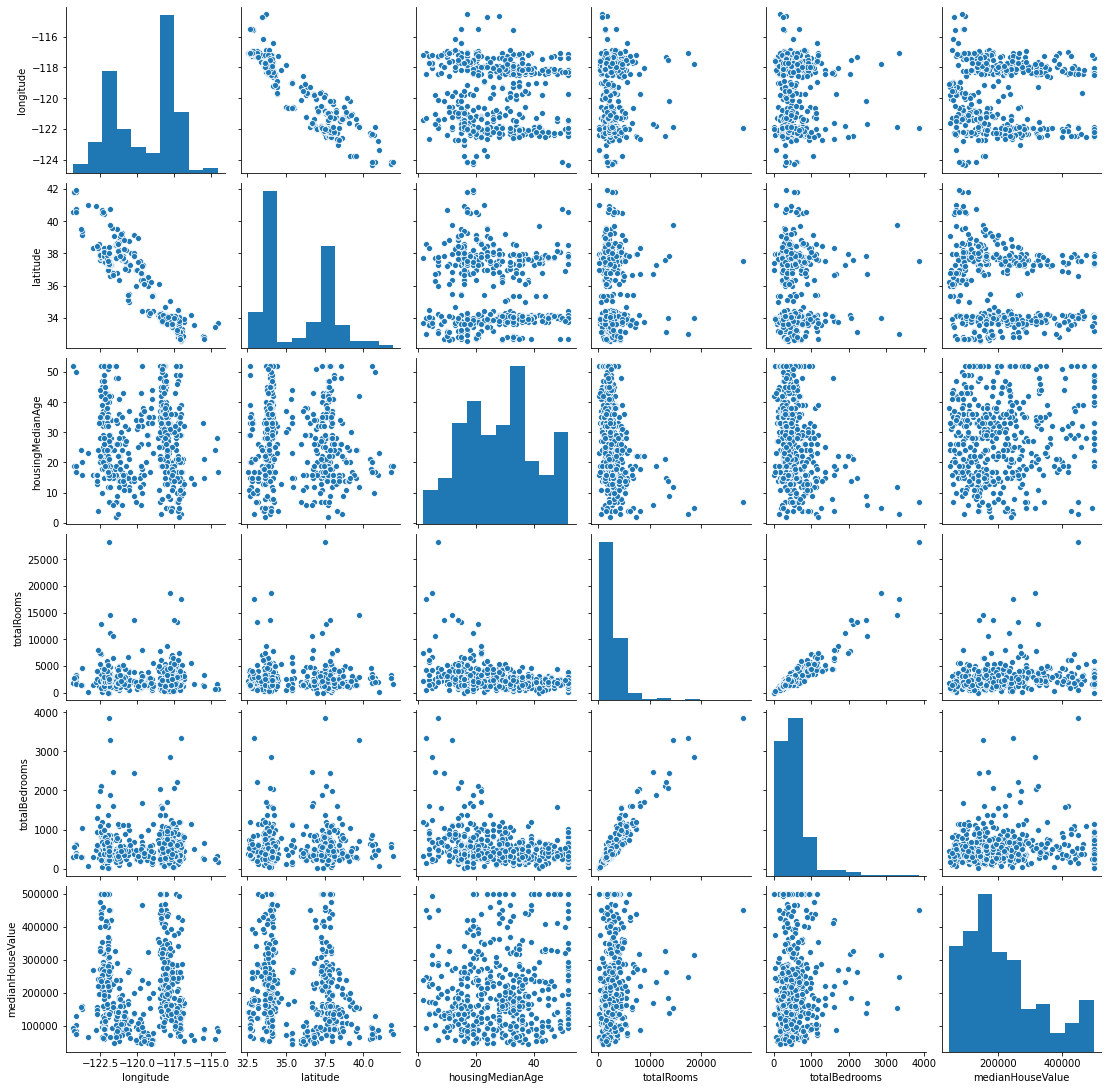
Understand data#
import seaborn as sns
g = sns.pairplot(d)
Show house locations on map#
observa como el valor de las casas es más caro en zonas urbanas
from bokeh.plotting import *
from bokeh.models import *
import bokeh
from matplotlib import cm
from sklearn.preprocessing import MinMaxScaler
def latlng_to_meters(lat, lng):
origin_shift = 2 * np.pi * 6378137 / 2.0
mx = lng * origin_shift / 180.0
my = np.log(np.tan((90 + lat) * np.pi / 360.0)) / (np.pi / 180.0)
my = my * origin_shift / 180.0
return mx, my
def xplot_map(lat, lon, color=None, size=10):
cmap = cm.rainbow
wlat, wlong = latlng_to_meters(lat, lon)
if color is not None:
colors = MinMaxScaler(feature_range=(0,255)).fit_transform(color)
colors = ["#%02x%02x%02x"%tuple([int(j*255) for j in cmap(int(i))[:3]]) for i in colors]
openmap_url = 'http://c.tile.openstreetmap.org/{Z}/{X}/{Y}.png'
otile_url = 'http://otile1.mqcdn.com/tiles/1.0.0/sat/{Z}/{X}/{Y}.jpg'
TILES = WMTSTileSource(url=openmap_url)
tools="pan,wheel_zoom,reset"
p = figure(tools=tools, min_width=700,min_height=600)
p.add_tile(TILES)
p.axis.visible = False
cb = figure(min_width=40, max_width=40, min_height=600, tools=tools)
yc = np.linspace(np.min(color),np.max(color),20)
c = np.linspace(0,255,20).astype(int)
dy = yc[1]-yc[0]
cb.rect(x=0.5, y=yc, color=["#%02x%02x%02x"%tuple([int(j*255) for j in cmap(int(i))[:3]]) for i in c], width=1, height = dy)
cb.xaxis.visible = False
p.circle(np.array(wlat), np.array(wlong), color=colors, size=size)
pb = gridplot([[p, cb]])
show(pb)
ds = d.sample(500)
xplot_map(ds["latitude"].values,
ds["longitude"].values, ds["medianHouseValue"].values.reshape(-1,1)/1e5)
Separate variable to predict#
X = d.values[:,:-1]
y = d["medianHouseValue"].values
print (X.shape, y.shape)
(500, 5) (500,)
from sklearn.linear_model import LinearRegression
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.metrics import median_absolute_error, r2_score, mean_squared_error
Xtr, Xts, ytr, yts = train_test_split(X,y, test_size=0.3)
print (Xtr.shape, ytr.shape, Xts.shape, yts.shape)
(350, 5) (350,) (150, 5) (150,)
A linear regression#
check sklearn LinearRegression doc to understand the
scorefunction.
lr = LinearRegression()
lr.fit(Xtr, ytr)
lr.score(Xtr, ytr), lr.score(Xts, yts)
(0.32232366147227454, 0.307618752063642)
r2_score(yts, lr.predict(Xts))
0.307618752063642
median_absolute_error(yts, lr.predict(Xts))
55453.23499294184
mean_squared_error(yts, lr.predict(Xts))
10000399171.317457
however we will create our score
mean releative absolute error
def rel_mrae(estimator, X, y):
preds = estimator.predict(X)
return np.mean(np.abs(preds-y)/y)
rel_mrae(lr, Xtr, ytr), rel_mrae(lr, Xts, yts)
(0.482339200932234, 0.3994545505967154)
let’s understand prediction errors
preds = lr.predict(Xts)
errors = np.abs(preds-yts)/yts
plt.figure(figsize=(20,3))
cols = ["longitude","latitude","housingMedianAge", "totalRooms","totalBedrooms"]
for i,col in enumerate(cols):
plt.subplot(1,len(cols),i+1)
plt.scatter(errors, Xts[:,i])
plt.ylabel(col)
plt.xlabel("relative error")
plt.grid();
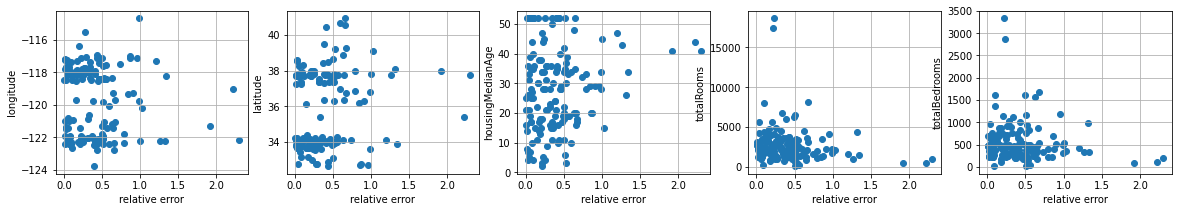
we observe there is no significant correlation between the error and any. It does seem that when the houseMedianAge is smaller, the error is also smaller, and when the totalBedrooms is higher the error is also smaller. However this seems to involve only a fraction of the houses. The correlation coefficients seems to capture this.
corrcoefs = pd.DataFrame([np.corrcoef(Xts[:,i], errors)[0,1] for i in range(len(cols))], index=cols, columns=["corrcoef"])
corrcoefs
| corrcoef | |
|---|---|
| longitude | -0.109087 |
| latitude | 0.172464 |
| housingMedianAge | 0.143974 |
| totalRooms | -0.140183 |
| totalBedrooms | -0.106749 |
How sure can we be of our model performance#
resample, train and measure
bootstrap: resample and put back
cross validation: resample and partition
from progressbar import progressbar as pbar
def bootstrap_score(estimator, X, y, test_size):
trscores, tsscores = [], []
for _ in range(10):
Xtr, Xts, ytr, yts = train_test_split(X,y, test_size=test_size)
estimator.fit(Xtr, ytr)
trscores.append(rel_mrae(estimator, Xtr, ytr))
tsscores.append(rel_mrae(estimator, Xts, yts))
return (np.mean(trscores), np.std(trscores)), (np.mean(tsscores), np.std(tsscores))
estimator = LinearRegression()
(trmean, trstd), (tsmean, tsstd) = bootstrap_score(estimator, X, y, test_size=0.3)
print ("train score %.3f (±%.4f)"%(trmean, trstd))
print ("test score %.3f (±%.4f)"%(tsmean, tsstd))
train score 0.469 (±0.0097)
test score 0.452 (±0.0255)
the sklearn library provides several validation methods
Bootstrapping: data is sampled randomly at every split
from sklearn.model_selection import ShuffleSplit, KFold,cross_val_score
ss = ShuffleSplit(n_splits=3, test_size=0.3)
for a,b in ss.split(range(10)):
print (a, b)
[6 5 1 8 4 7 0] [9 3 2]
[4 7 8 3 6 0 1] [2 9 5]
[1 0 7 4 9 5 6] [8 2 3]
z = cross_val_score(lr, X, y, cv = ShuffleSplit(n_splits=10, test_size=0.3), scoring=rel_mrae)
print (z)
print ("test score %.3f (±%.4f)"%(np.mean(z), np.std(z)))
[0.47475243 0.4532451 0.45386072 0.47604051 0.48315741 0.57676936
0.45063058 0.48636779 0.47881854 0.46195466]
test score 0.480 (±0.0347)
Cross Validation: data is partitioned
ss = KFold(n_splits=3)
for a,b in ss.split(range(10)):
print (a, b)
[4 5 6 7 8 9] [0 1 2 3]
[0 1 2 3 7 8 9] [4 5 6]
[0 1 2 3 4 5 6] [7 8 9]
z = cross_val_score(lr, X, y, cv = KFold(n_splits=10), scoring=rel_mrae)
print (z)
print ("test score %.3f (±%.4f)"%(np.mean(z), np.std(z)))
[0.34715741 0.56105641 0.46435799 0.45752588 0.49913672 0.40221641
0.50140556 0.41300136 0.61422836 0.45926731]
test score 0.472 (±0.0736)
assess the score with a learning curve
from sklearn.model_selection import ShuffleSplit
cv = ShuffleSplit(n_splits=10, test_size=.3)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X, y,
cv=cv, scoring=rel_mrae, ylim=(0,0.7))
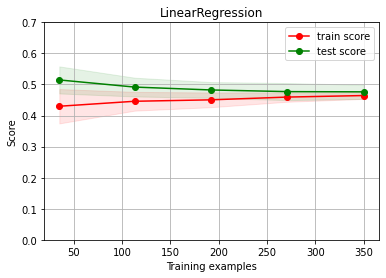
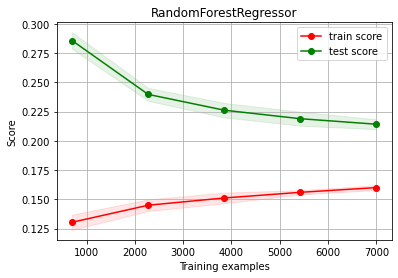
Diagnosing#
Linear regression BASELINE
estimator = LinearRegression()
cv = ShuffleSplit(n_splits=10, test_size=.3)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X, y, cv=cv, scoring=rel_mrae, ylim=(0,0.7))
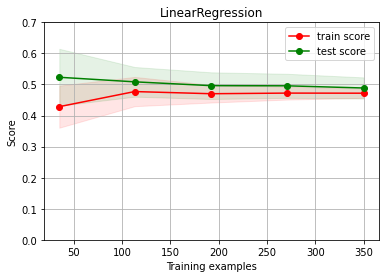
We have UNDERFITTING (high bias)
increase model complexity
get more columns
# try first increasing model complexity --> a bit better but with overfitting
# experiment with different max_depth values
from sklearn.tree import DecisionTreeRegressor
estimator = DecisionTreeRegressor(max_depth=8)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X, y, cv=cv, scoring=rel_mrae, ylim=(0,0.7))
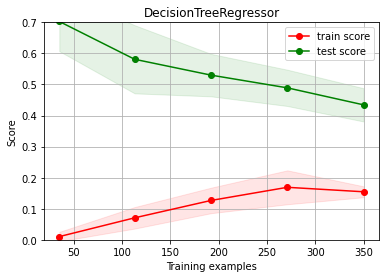
# try now with more columns (we have them!!!) --> improves a bit
d2 = pd.read_csv("local/data/cal_housing_small_full.data")
d2.head()
| longitude | latitude | housingMedianAge | totalRooms | totalBedrooms | medianHouseValue | population | households | medianIncome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -120.58 | 35.00 | 37.0 | 523.0 | 119.0 | 106300.0 | 374.0 | 95.0 | 1.4726 |
| 1 | -118.17 | 33.98 | 31.0 | 1236.0 | 329.0 | 155400.0 | 1486.0 | 337.0 | 3.0938 |
| 2 | -122.22 | 37.81 | 52.0 | 1971.0 | 335.0 | 273700.0 | 765.0 | 308.0 | 6.5217 |
| 3 | -117.91 | 33.66 | 21.0 | 1708.0 | 505.0 | 193800.0 | 1099.0 | 434.0 | 3.2250 |
| 4 | -121.92 | 37.24 | 27.0 | 1265.0 | 216.0 | 281200.0 | 660.0 | 232.0 | 5.3911 |
estimator = LinearRegression()
X2 = d2[[col for col in d2.columns if col!="medianHouseValue"]].values
y2 = d2["medianHouseValue"].values
print (X2.shape, y2.shape)
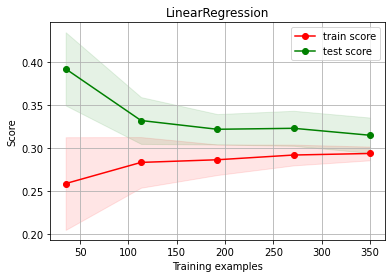
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X2, y2, cv=cv, scoring=rel_mrae)
(500, 8) (500,)
Diagnosing#
Random Forest BASELINE
from sklearn.ensemble import RandomForestRegressor
estimator = RandomForestRegressor(max_depth=10)
(trmean, trstd), (tsmean, tsstd) = bootstrap_score(estimator, X, y, test_size=0.3)
print ("train score %.3f (±%.4f)"%(trmean, trstd))
print ("test score %.3f (±%.4f)"%(tsmean, tsstd))
train score 0.158 (±0.0078)
test score 0.362 (±0.0226)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X, y, cv=cv, scoring=rel_mrae)
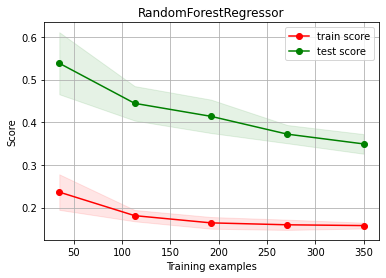
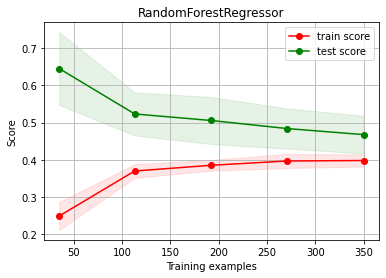
We have OVERFITTING (high variance)
reduce model complexity
get more data
# try first reduce model complexity --> more BIAS
estimator = RandomForestRegressor(max_depth=4)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X, y, cv=cv, scoring=rel_mrae)
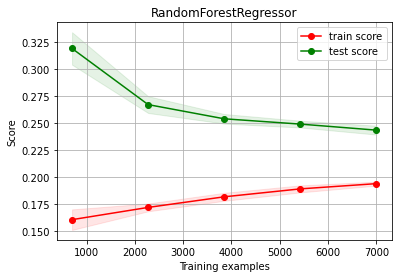
# try now with more data (we have A LOT!!!)
d3 = pd.read_csv("local/data/cal_housing.data")
print ("TOTAL AVAILABLE DATA", d3.shape)
d3 = d3.sample(10000)
estimator = RandomForestRegressor(max_depth=10)
X3 = d3.values[:,:-1]
y3 = d3["medianHouseValue"].values
print ("building learning curve with", X3.shape, y3.shape)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X3, y3, cv=cv, scoring=rel_mrae)
TOTAL AVAILABLE DATA (20640, 6)
building learning curve with (10000, 5) (10000,)
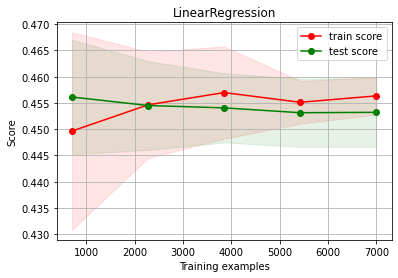
What if we made the wrong choice#
Linear Regression UNDERFITTING and choose acquire more data: No improvement!!!
estimator = LinearRegression()
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X3, y3, cv=cv, scoring=rel_mrae)
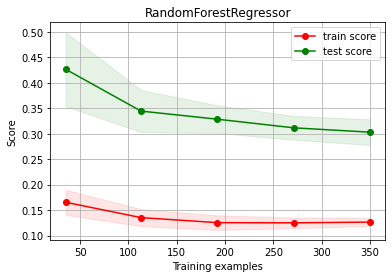
Random Forest OVERFITTING and choose to add more columns: some improvement!!!
estimator = RandomForestRegressor(max_depth=10)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X2, y2, cv=cv, scoring=rel_mrae)
let’s now choose more data and more columns (a luxury!!!)
d4 = pd.read_csv("local/data/cal_housing_full.data")
print ("TOTAL AVAILABLE DATA", d4.shape)
d4 = d4.sample(10000)
estimator = RandomForestRegressor(max_depth=10)
X4 = d4.values[:,:-1]
y4 = d4["medianHouseValue"].values
print ("building learning curve with", X4.shape, y4.shape)
ch.plot_learning_curve(estimator, estimator.__class__.__name__, X4, y4, cv=cv, scoring=rel_mrae)
TOTAL AVAILABLE DATA (20640, 9)
building learning curve with (10000, 8) (10000,)